4BKT
 
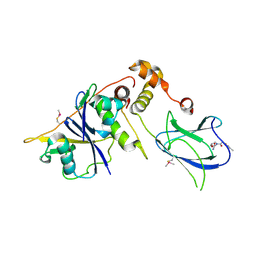 | | von Hippel Lindau protein:ElonginB:ElonginC complex, in complex with (2S,4R)-N-methyl-1-[2-(3-methyl-1,2-oxazol-5-yl)ethanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide | | Descriptor: | (2S,4R)-N-methyl-1-[2-(3-methyl-1,2-oxazol-5-yl)ethanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, ARGININE, GLUTAMIC ACID, ... | | Authors: | Van Molle, I, Dias, D.M, Baud, M, Galdeano, C, Geraldes, C.F.G.C, Ciulli, A. | | Deposit date: | 2013-04-29 | | Release date: | 2013-11-27 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Is NMR Fragment Screening Fine-Tuned to Assess Druggability of Protein-Protein Interactions?
Acs Med.Chem.Lett., 5, 2014
|
|
7LLI
 
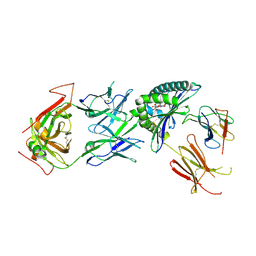 | | Stimulatory immune receptor protein complex | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, Major histocompatibility complex class I-related gene protein, ... | | Authors: | Rice, M.T, Littler, D.R, Rossjohn, J, Gully, B.S. | | Deposit date: | 2021-02-04 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Recognition of the antigen-presenting molecule MR1 by a V delta 3 + gamma delta T cell receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LLJ
 
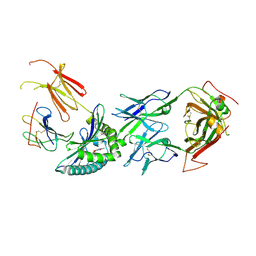 | | Inhibitory immune receptor protein complex | | Descriptor: | Beta-2-microglobulin, Major histocompatibility complex class I-related gene protein, N-(6-formyl-4-oxo-3,4-dihydropteridin-2-yl)acetamide, ... | | Authors: | Rice, M.T, Littler, D.R, Rossjohn, J, Gully, B.S. | | Deposit date: | 2021-02-04 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Recognition of the antigen-presenting molecule MR1 by a V delta 3 + gamma delta T cell receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6Q7H
 
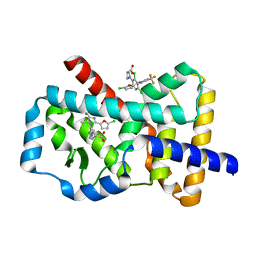 | |
8F6Y
 
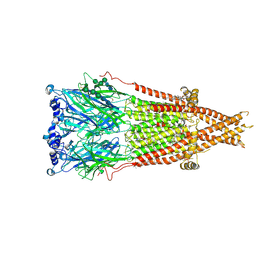 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with etomidate, desensitized-like state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goswami, U, Rahman, M.M, Teng, J, Hibbs, R.E. | | Deposit date: | 2022-11-17 | | Release date: | 2023-06-07 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural interplay of anesthetics and paralytics on muscle nicotinic receptors.
Nat Commun, 14, 2023
|
|
1ELZ
 
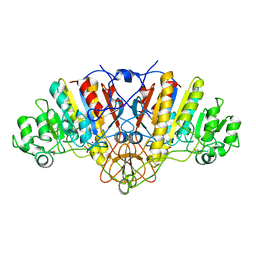 | | E. COLI ALKALINE PHOSPHATASE MUTANT (S102G) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Stec, B, Hehir, M, Brennan, C, Nolte, M, Kantrowitz, E.R. | | Deposit date: | 1998-02-10 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Kinetic and X-ray structural studies of three mutant E. coli alkaline phosphatases: insights into the catalytic mechanism without the nucleophile Ser102.
J.Mol.Biol., 277, 1998
|
|
1RNQ
 
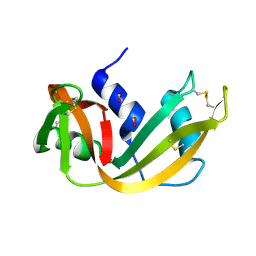 | | RIBONUCLEASE A CRYSTALLIZED FROM 8M SODIUM FORMATE | | Descriptor: | FORMIC ACID, RIBONUCLEASE A | | Authors: | Fedorov, A.A, Josef-Mccarthy, D, Graf, I, Anguelova, D, Fedorov, E.V, Almo, S.C. | | Deposit date: | 1995-11-08 | | Release date: | 1996-04-03 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ionic interactions in crystalline bovine pancreatic ribonuclease A.
Biochemistry, 35, 1996
|
|
7BBF
 
 | |
8ESK
 
 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with rocuronium, resting-like state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goswami, U, Rahman, M.M, Teng, J, Hibbs, R.E. | | Deposit date: | 2022-10-14 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural interplay of anesthetics and paralytics on muscle nicotinic receptors.
Nat Commun, 14, 2023
|
|
8F6Z
 
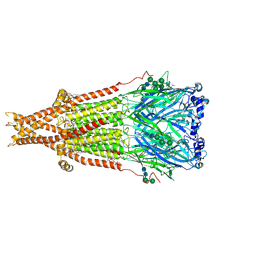 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with succinylcholine, desensitized-like state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2,2'-[(1,4-DIOXOBUTANE-1,4-DIYL)BIS(OXY)]BIS(N,N,N-TRIMETHYLETHANAMINIUM), 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goswami, U, Rahman, M.M, Teng, J, Hibbs, R.E. | | Deposit date: | 2022-11-17 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural interplay of anesthetics and paralytics on muscle nicotinic receptors.
Nat Commun, 14, 2023
|
|
8F2S
 
 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with rocuronium, pore-blocked state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goswami, U, Rahman, M.M, Teng, J, Hibbs, R.E. | | Deposit date: | 2022-11-08 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural interplay of anesthetics and paralytics on muscle nicotinic receptors.
Nat Commun, 14, 2023
|
|
7NQ8
 
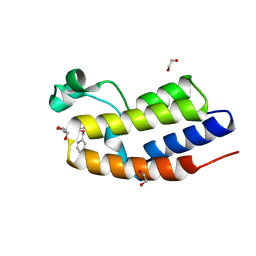 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH N-ethyl-3-(1-methyl-1H-1,2,3-triazol-4-yl)-4-(pyridin-2-ylmethoxy)benzamide | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, N-ethyl-3-(1-methyl-1H-1,2,3-triazol-4-yl)-4-(pyridin-2-ylmethoxy)benzamide | | Authors: | Chung, C.W. | | Deposit date: | 2021-03-01 | | Release date: | 2021-03-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Template-Hopping Approach Leads to Potent, Selective, and Highly Soluble Bromo and Extraterminal Domain (BET) Second Bromodomain (BD2) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7NQI
 
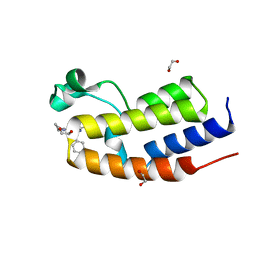 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 2-benzyl-N-cyclopropyl-6-(1-methyl-1H-1,2,3-triazol-4-yl)isonicotinamide | | Descriptor: | 1,2-ETHANEDIOL, 2-benzyl-N-cyclopropyl-6-(1-methyl-1H-1,2,3-triazol-4-yl)isonicotinamide, Bromodomain-containing protein 2 | | Authors: | Chung, C.W. | | Deposit date: | 2021-03-01 | | Release date: | 2021-03-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Template-Hopping Approach Leads to Potent, Selective, and Highly Soluble Bromo and Extraterminal Domain (BET) Second Bromodomain (BD2) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7NQ9
 
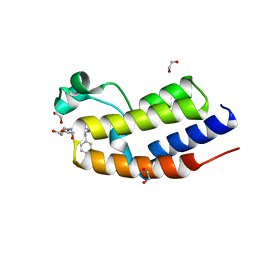 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 2-benzyl-N-cyclopropyl-6-(1-methyl-1H-1,2,3-triazol-4-yl)isonicotinamide | | Descriptor: | 1,2-ETHANEDIOL, 6-benzyl-N4-((1r,3r)-3-hydroxycyclobutyl)-N2-methylpyridine-2,4-dicarboxamide, Bromodomain-containing protein 2 | | Authors: | Chung, C.W. | | Deposit date: | 2021-03-01 | | Release date: | 2021-03-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Template-Hopping Approach Leads to Potent, Selective, and Highly Soluble Bromo and Extraterminal Domain (BET) Second Bromodomain (BD2) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7NQJ
 
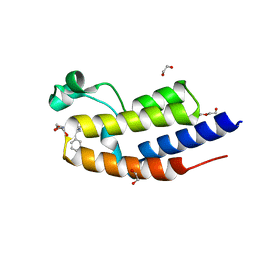 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH N-ethyl-2-(1-methyl-1H-1,2,3-triazol-4-yl)-6-(1-phenylethyl)isonicotinamide | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chung, C. | | Deposit date: | 2021-03-01 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.734 Å) | | Cite: | Template-Hopping Approach Leads to Potent, Selective, and Highly Soluble Bromo and Extraterminal Domain (BET) Second Bromodomain (BD2) Inhibitors.
J.Med.Chem., 64, 2021
|
|
5HUT
 
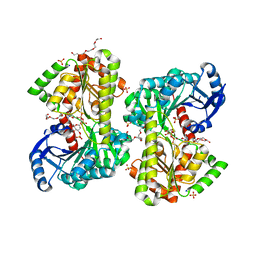 | | Structure of Candida albicans trehalose-6-phosphate synthase in complex with UDP-glucose | | Descriptor: | Alpha,alpha-trehalose-phosphate synthase [UDP-forming], PENTAETHYLENE GLYCOL, SODIUM ION, ... | | Authors: | Miao, Y, Brennan, R.G. | | Deposit date: | 2016-01-27 | | Release date: | 2017-05-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and In Vivo Studies on Trehalose-6-Phosphate Synthase from Pathogenic Fungi Provide Insights into Its Catalytic Mechanism, Biological Necessity, and Potential for Novel Antifungal Drug Design.
MBio, 8, 2017
|
|
1VFH
 
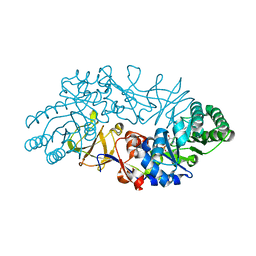 | | Crystal structure of alanine racemase from D-cycloserine producing Streptomyces lavendulae | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, alanine racemase | | Authors: | Noda, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2004-04-13 | | Release date: | 2004-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence that alanine racemase from a D-cycloserine-producing microorganism exhibits resistance to its own product.
J.Biol.Chem., 279, 2004
|
|
1VFS
 
 | | Crystal structure of D-cycloserine-bound form of alanine racemase from D-cycloserine-producing Streptomyces lavendulae | | Descriptor: | CHLORIDE ION, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, alanine racemase | | Authors: | Noda, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2004-04-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural evidence that alanine racemase from a D-cycloserine-producing microorganism exhibits resistance to its own product.
J.Biol.Chem., 279, 2004
|
|
4A2E
 
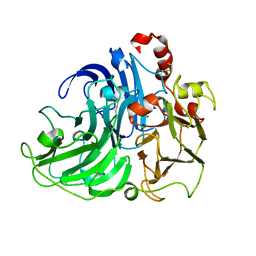 | | Crystal Structure of a Coriolopsis gallica Laccase at 1.7 A Resolution pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, LACCASE | | Authors: | De La Mora, E, Valderrama, B, Horjales, E, Rudino-Pinera, E. | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Changes Caused by Radiation-Induced Reduction and Radiolysis: The Effect of X-Ray Absorbed Dose in a Fungal Multicopper Oxidase
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1RIT
 
 | | Crystal structure of Peanut lectin in complex with meso-tetrasulphonatophenylporphyrin and lactose | | Descriptor: | 5,10,15,20-TETRAKIS(4-SULPFONATOPHENYL)-21H,23H-PORPHINE, CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Goel, M, Kaur, K.J, Maiya, B.G, Swamy, M.J, Salunke, D.M. | | Deposit date: | 2003-11-17 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structures of the PNA-porphyrin complex in the presence and absence of lactose: mapping the conformational changes on lactose binding, interacting surfaces, and supramolecular aggregations.
Biochemistry, 44, 2005
|
|
7WMI
 
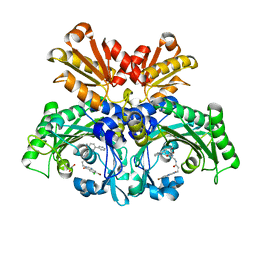 | | Threonyl-tRNA synthetase from Salmonella enterica in complex with an inhibitor | | Descriptor: | (2S,3R)-2-azanyl-N-[(1R)-2-[3-(7-bromanyl-6-chloranyl-4-oxidanylidene-quinazolin-3-yl)propanoylamino]-1-(3-phenylphenyl)ethyl]-3-oxidanyl-butanamide, 1,2-ETHANEDIOL, Threonine--tRNA ligase, ... | | Authors: | Cai, Z, Chen, B, Yu, Y, Zhou, H. | | Deposit date: | 2022-01-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Proof-of-Concept of Triple-Site Inhibitors against Aminoacyl-tRNA Synthetases.
J.Med.Chem., 65, 2022
|
|
6VXG
 
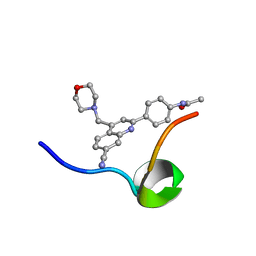 | | Structure of the C-terminal Domain of RAGE and Its Inhibitor | | Descriptor: | Advanced glycosylation end product-specific receptor, N-(4-{7-cyano-4-[(morpholin-4-yl)methyl]quinolin-2-yl}phenyl)acetamide | | Authors: | Ramirez, L, Shekhtman, A. | | Deposit date: | 2020-02-21 | | Release date: | 2021-02-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Small-molecule antagonism of the interaction of the RAGE cytoplasmic domain with DIAPH1 reduces diabetic complications in mice.
Sci Transl Med, 13, 2021
|
|
4TXN
 
 | | Crystal structure of uridine phosphorylase from Schistosoma mansoni in complex with 5-fluorouracil | | Descriptor: | 5-FLUOROURACIL, SULFATE ION, Uridine phosphorylase | | Authors: | Marinho, A, Torini, J, Romanello, L, Cassago, A, DeMarco, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2014-07-03 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of two Schistosoma mansoni uridine phosphorylases isoforms suggests the emergence of a protein with a non-canonical function.
Biochimie, 125, 2016
|
|
4CEV
 
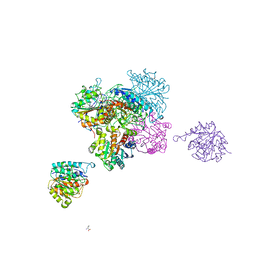 | | ARGINASE FROM BACILLUS CALDEVELOX, L-ORNITHINE COMPLEX | | Descriptor: | GUANIDINE, L-ornithine, MANGANESE (II) ION, ... | | Authors: | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | Deposit date: | 1999-03-15 | | Release date: | 1999-04-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
8BYA
 
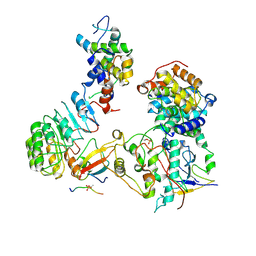 | | Cryo-EM structure of SKP1-SKP2-CKS1-CDK2-CyclinA-p27KIP1 Complex | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, Cyclin-dependent kinase inhibitor 1B, ... | | Authors: | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
