2SOC
 
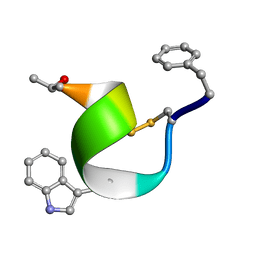 | |
5IPE
 
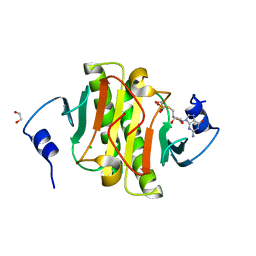 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) nucleoside thiophosphoramidate catalytic product complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-phosphono-5'-thioguanosine, CHLORIDE ION, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Caught before Released: Structural Mapping of the Reaction Trajectory for the Sofosbuvir Activating Enzyme, Human Histidine Triad Nucleotide Binding Protein 1 (hHint1).
Biochemistry, 56, 2017
|
|
5IPC
 
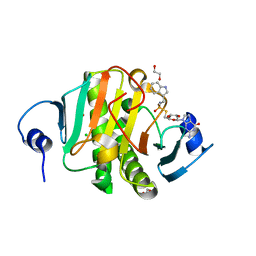 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside thiophosphoramidate substrate complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-[(S)-hydroxy{[2-(1H-indol-3-yl)ethyl]amino}phosphoryl]-5'-thioguanosine, CHLORIDE ION, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Caught before Released: Structural Mapping of the Reaction Trajectory for the Sofosbuvir Activating Enzyme, Human Histidine Triad Nucleotide Binding Protein 1 (hHint1).
Biochemistry, 56, 2017
|
|
5IPD
 
 | |
2VKA
 
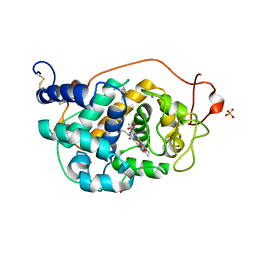 | | Site-Directed Mutagenesis of the Catalytic Tryptophan Environment in Pleurotus eryngii Versatile Peroxidase | | Descriptor: | CALCIUM ION, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ruiz-Duenas, F.J, Morales, M, Mate, M.J, Romero, A, Martinez, M.J, Smith, A, Martinez, A.T. | | Deposit date: | 2007-12-18 | | Release date: | 2008-01-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Site-Directed Mutagenesis of the Catalytic Tryptophan Environment in Pleurotus Eryngii Versatile Peroxidase
Biochemistry, 47, 2008
|
|
3WNZ
 
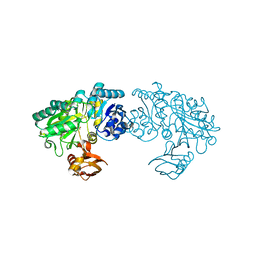 | | Crystal structure of Bacillus subtilis YwfE, an L-amino acid ligase, with bound ADP-Mg-Pi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alanine-anticapsin ligase BacD, MAGNESIUM ION, ... | | Authors: | Tsuda, T, Kojima, S. | | Deposit date: | 2013-12-19 | | Release date: | 2014-05-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Single Mutation Alters the Substrate Specificity of l-Amino Acid Ligase
Biochemistry, 53, 2014
|
|
2D2O
 
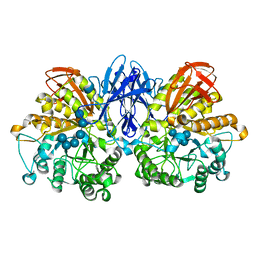 | | Structure of a complex of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with maltohexaose demonstrates the important role of aromatic residues at the reducing end of the substrate binding cleft | | Descriptor: | CALCIUM ION, Neopullulanase 2, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ohtaki, A, Mizuno, M, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-09-13 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a complex of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with maltohexaose demonstrates the important role of aromatic residues at the reducing end of the substrate binding cleft
Carbohydr.Res., 341, 2006
|
|
3WO0
 
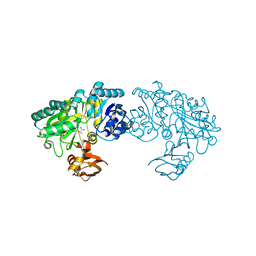 | | Crystal structure of Bacillus subtilis YwfE, an L-amino acid ligase, with bound ADP-Mg-Ala | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALANINE, Alanine-anticapsin ligase BacD, ... | | Authors: | Tsuda, T, Kojima, S. | | Deposit date: | 2013-12-19 | | Release date: | 2014-05-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single Mutation Alters the Substrate Specificity of l-Amino Acid Ligase
Biochemistry, 53, 2014
|
|
2J44
 
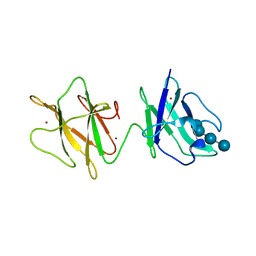 | | Alpha-glucan binding by a streptococcal virulence factor | | Descriptor: | ALKALINE AMYLOPULLULANASE, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Lammerts van Bueren, A, Higgins, M, Wang, D, Burke, R.D, Boraston, A.B. | | Deposit date: | 2006-08-24 | | Release date: | 2006-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and Structural Basis of Binding to Host Lung Glycogen by Streptococcal Virulence Factors.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1QZ9
 
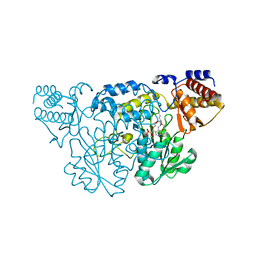 | | The Three Dimensional Structure of Kynureninase from Pseudomonas fluorescens | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECANE, CHLORIDE ION, KYNURENINASE, ... | | Authors: | Momany, C, Levdikov, V, Blagova, L, Lima, S, Phillips, R.S. | | Deposit date: | 2003-09-16 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-Dimensional Structure of Kynureninase from Pseudomonas fluorescens.
Biochemistry, 43, 2004
|
|
1QMT
 
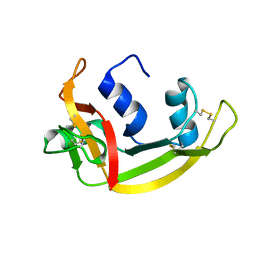 | |
5CHY
 
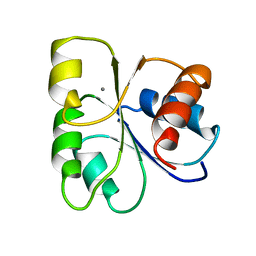 | | STRUCTURE OF CHEMOTAXIS PROTEIN CHEY | | Descriptor: | CALCIUM ION, CHEY | | Authors: | Zhu, X, Rebello, J, Matsumura, P, Volz, K. | | Deposit date: | 1996-08-29 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of CheY mutants Y106W and T87I/Y106W. CheY activation correlates with movement of residue 106.
J.Biol.Chem., 272, 1997
|
|
1IIL
 
 | | CRYSTAL STRUCTURE OF PRO253ARG APERT MUTANT FGF RECEPTOR 2 (FGFR2) IN COMPLEX WITH FGF2 | | Descriptor: | FIBROBLAST GROWTH FACTOR RECEPTOR 2, HEPARIN-BINDING GROWTH FACTOR 2 | | Authors: | Ibrahimi, O.A, Eliseenkova, A.V, Plotnikov, A.N, Ornitz, D.M, Mohammadi, M. | | Deposit date: | 2001-04-23 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for fibroblast growth factor receptor 2 activation in Apert syndrome.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1OKL
 
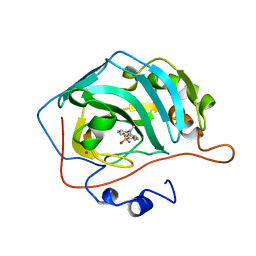 | | CARBONIC ANHYDRASE II COMPLEX WITH THE 1OKL INHIBITOR 5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONAMIDE | | Descriptor: | 5-(DIMETHYLAMINO)-1-NAPHTHALENESULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Elbaum, D, Christianson, D.W. | | Deposit date: | 1996-06-25 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected binding mode of the sulfonamide fluorophore 5-dimethylamino-1-naphthalene sulfonamide to human carbonic anhydrase II. Implications for the development of a zinc biosensor.
J.Biol.Chem., 271, 1996
|
|
1H41
 
 | | Pseudomonas cellulosa E292A alpha-D-glucuronidase mutant complexed with aldotriuronic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-O-methyl-alpha-D-glucopyranuronic acid, ALPHA-GLUCURONIDASE, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-09-25 | | Release date: | 2003-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Alpha-Glucuronidase,Glca67A,of Cellvibrio Japonicus Utilizes the Carboxylate and Methyl Groups of Aldobiouronic Acid as Important Substrate Recognition Determinants
J.Biol.Chem., 278, 2003
|
|
7CUE
 
 | | Crystal structure of HID2 bound to human Hemoglobin | | Descriptor: | Amino acid ABC transporter substrate-binding protein, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Caaveiro, J.M.M, Hoshino, M, Tsumoto, K. | | Deposit date: | 2020-08-22 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the recognition of human hemoglobin by the Shr protein from Streptococcus pyogenes
To Be Published
|
|
1IMI
 
 | | SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 | | Descriptor: | PROTEIN (ALPHA-CONOTOXIN IMI) | | Authors: | Maslennikov, I.V, Shenkarev, Z.O, Zhmak, M.N, Tsetlin, V.I, Ivanov, V.T, Arseniev, A.S. | | Deposit date: | 1998-11-27 | | Release date: | 1999-04-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR spatial structure of alpha-conotoxin ImI reveals a common scaffold in snail and snake toxins recognizing neuronal nicotinic acetylcholine receptors.
FEBS Lett., 444, 1999
|
|
1HWH
 
 | |
1S6V
 
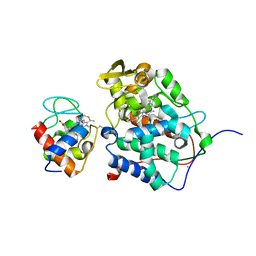 | | Structure of a cytochrome c peroxidase-cytochrome c site specific cross-link | | Descriptor: | Cytochrome c peroxidase, mitochondrial, Cytochrome c, ... | | Authors: | Guo, M, Bhaskar, B, Li, H, Barrows, T.P, Poulos, T.L. | | Deposit date: | 2004-01-27 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure and characterization of a cytochrome c peroxidase-cytochrome c site-specific cross-link
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2EVQ
 
 | |
1HWG
 
 | |
8P2W
 
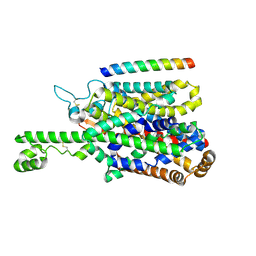 | | Structure of human SIT1 (focussed map / refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Sodium- and chloride-dependent transporter XTRP3 | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P2Z
 
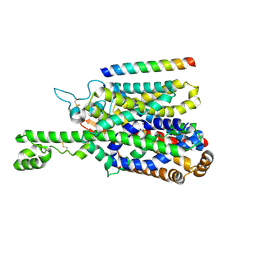 | | Structure of human SIT1 bound to L-pipecolate (focussed map / refinement) | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P30
 
 | | Structure of human SIT1:ACE2 complex (open PD conformation) bound to L-pipecolate | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P2Y
 
 | | Structure of human SIT1:ACE2 complex (closed PD conformation) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
