8UKN
 
 | | APO and AMP-PNP bound cAMP-dependent protein kinase A catalytic domain | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2023-10-14 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystallographic, kinetic, and calorimetric investigation of PKA interactions with L-type calcium channels and Rad GTPase.
J.Biol.Chem., 301, 2024
|
|
8GSU
 
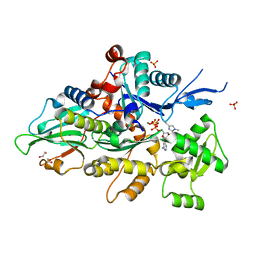 | | Crystal structure of human cardiac alpha actin (WT_ADP-Pi) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
8GT4
 
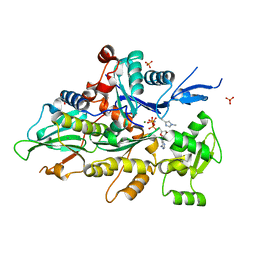 | | Crystal structure of human cardiac alpha actin Q137A mutant (AMPPNP state) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, Actin, alpha cardiac muscle 1, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
8GT1
 
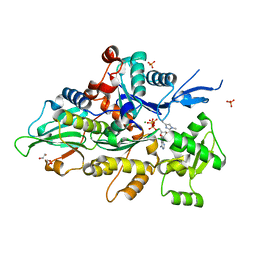 | | Crystal structure of human cardiac alpha actin A108G mutant (ADP-Pi state) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
8GT5
 
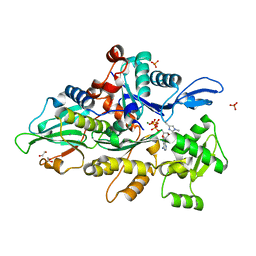 | | Crystal structure of human cardiac alpha actin Q137A mutant (ADP-Pi state) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
8GSW
 
 | | Crystal structure of human cardiac alpha actin A108G mutant (AMPPNP state) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, Actin, alpha cardiac muscle 1, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
8GT2
 
 | | Crystal structure of human cardiac alpha actin P109A mutant (AMPPNP state) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, Actin, alpha cardiac muscle 1, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
8GT3
 
 | | Crystal structure of human cardiac alpha actin P109A mutant (ADP-Pi state) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
4TN3
 
 | | Structure of the BBox-Coiled-coil region of Rhesus Trim5alpha | | Descriptor: | TRIM5/cyclophilin A fusion protein/T4 Lysozyme chimera, ZINC ION | | Authors: | Kirkpatrick, J.J, Stoye, J.P, Taylor, I.A, Goldstone, D.C. | | Deposit date: | 2014-06-03 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1989 Å) | | Cite: | Structural studies of postentry restriction factors reveal antiparallel dimers that enable avid binding to the HIV-1 capsid lattice.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8FQQ
 
 | | ADC-162 in complex with boronic acid transition state inhibitor MB076 | | Descriptor: | 1-[(2R)-2-{2-[(5-amino-1,3,4-thiadiazol-2-yl)sulfanyl]acetamido}-2-boronoethyl]-1H-1,2,3-triazole-4-carboxylic acid, Beta-lactamase, GLYCINE, ... | | Authors: | Powers, R.A, Wallar, B.J, June, C.M, Fernando, M.C. | | Deposit date: | 2023-01-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Synthesis of a Novel Boronic Acid Transition State Inhibitor, MB076: A Heterocyclic Triazole Effectively Inhibits Acinetobacter -Derived Cephalosporinase Variants with an Expanded-Substrate Spectrum.
J.Med.Chem., 66, 2023
|
|
7NY7
 
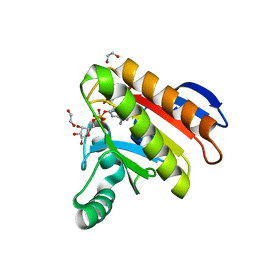 | | Crystal structure of the Capsaspora owczarzaki macroH2A macrodomain in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Histone macroH2A1.1 | | Authors: | Guberovic, I, Knobloch, G, Basquin, J, Buschbeck, M, Ladurner, A.G. | | Deposit date: | 2021-03-21 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of a histone variant involved in compartmental regulation of NAD metabolism.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7Q3A
 
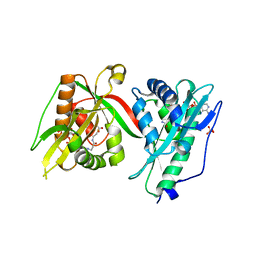 | | Crystal structure of MAB_4324 a tandem repeat GNAT from Mycobacterium abscessus | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Blaise, M, Alsarraf, M.A.B. | | Deposit date: | 2021-10-27 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical, structural, and functional studies reveal that MAB_4324c from Mycobacterium abscessus is an active tandem repeat N-acetyltransferase.
Febs Lett., 596, 2022
|
|
4S27
 
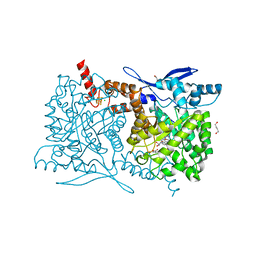 | | Crystal structure of Arabidopsis thaliana ThiC with bound aminoimidazole ribonucleotide, 5'-deoxyadenosine, L-methionine, Fe4S4 cluster and Fe | | Descriptor: | 1,4-BUTANEDIOL, 5'-DEOXYADENOSINE, 5-AMINOIMIDAZOLE RIBONUCLEOTIDE, ... | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
4SLI
 
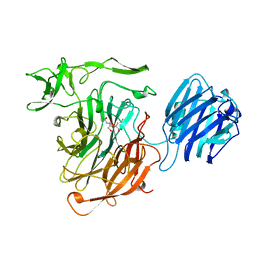 | | LEECH INTRAMOLECULAR TRANS-SIALIDASE COMPLEXED WITH 2-PROPENYL-NEU5AC, AN INACTIVE SUBSTRATE ANALOGUE | | Descriptor: | 2-propenyl-N-acetyl-neuraminic acid, INTRAMOLECULAR TRANS-SIALIDASE | | Authors: | Luo, Y, Li, S.C, Li, Y.T, Luo, M. | | Deposit date: | 1998-10-04 | | Release date: | 1999-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A structures of leech intramolecular trans-sialidase complexes: evidence of its enzymatic mechanism.
J.Mol.Biol., 285, 1999
|
|
4X11
 
 | | JC Polyomavirus genotype 3 VP1 in complex with GD1a oligosaccharide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Major capsid protein, ... | | Authors: | Stroh, L.J, Stehle, T. | | Deposit date: | 2014-11-24 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Greater Affinity of JC Polyomavirus Capsid for alpha 2,6-Linked Lactoseries Tetrasaccharide c than for Other Sialylated Glycans Is a Major Determinant of Infectivity.
J.Virol., 89, 2015
|
|
4RMD
 
 | | Crystal structure of human Retinoid X receptor alpha ligand binding domain complex with 9cUAB110 and coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-8-[3-cyclopropyl-2-(3-methylbutyl)cyclohex-2-en-1-ylidene]-3,7-dimethylocta-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Muccio, D.D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformationally Defined Rexinoids and Their Efficacy in the Prevention of Mammary Cancers.
J.Med.Chem., 58, 2015
|
|
6XDK
 
 | |
8W4Q
 
 | | Crystal structure of PDE4D complexed with CX-4945 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
7Q00
 
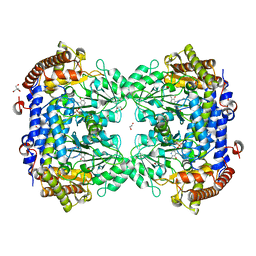 | | Crystal structure of serine hydroxymethyltransferase, isoform 4 from Arabidopsis thaliana (SHM4) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Serine hydroxymethyltransferase 4 | | Authors: | Ruszkowski, M, Sekula, B. | | Deposit date: | 2021-10-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Arabidopsis thaliana serine hydroxymethyltransferases: functions, structures, and perspectives.
Plant Physiol Biochem., 187, 2022
|
|
4XC1
 
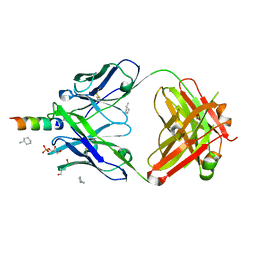 | | Crystal structure of human 4E10 Fab in complex with its peptide epitope on HIV-1 GP41: crystals cryoprotected with sn-Glycerol 3-phosphate | | Descriptor: | 4E10 FAB HEAVY CHAIN, 4E10 FAB LIGHT CHAIN, CYCLOHEXYLAMMONIUM ION, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
4RMC
 
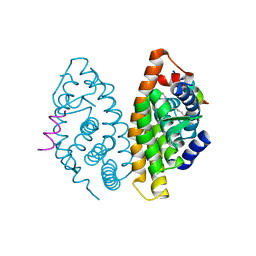 | | Crystal Structure of human retinoid X receptor alpha-ligand binding domain complex with 9cUAB76 and the coactivator peptide GRIP-1 | | Descriptor: | (3S,7S,8E)-8-[3-ethyl-2-(3-methylbutyl)cyclohex-2-en-1-ylidene]-3,7-dimethyloctanoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Muccio, D.D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformationally Defined Rexinoids and Their Efficacy in the Prevention of Mammary Cancers.
J.Med.Chem., 58, 2015
|
|
4XCP
 
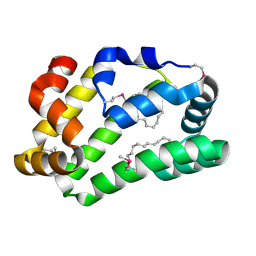 | | Fatty Acid and Retinol binding protein Na-FAR-1 from Necator americanus | | Descriptor: | Nematode fatty acid retinoid binding protein, PALMITIC ACID | | Authors: | Gabrielsen, M, Rey-Burusco, M.F, Ibanez-Shimabukuro, M, Griffiths, K, Kennedy, M.W, Corsico, B, Smith, B.O. | | Deposit date: | 2014-12-18 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Diversity in the structures and ligand-binding sites of nematode fatty acid and retinol-binding proteins revealed by Na-FAR-1 from Necator americanus.
Biochem.J., 471, 2015
|
|
4RPT
 
 | |
6XW7
 
 | | Crystal structure of murine norovirus P domain in complex with Nanobody NB-5829 and glycochenodeoxycholate (GCDCA) | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, GLYCOCHENODEOXYCHOLIC ACID, ... | | Authors: | Kilic, T, Sabin, C, Hansman, G. | | Deposit date: | 2020-01-23 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Nanobody-Mediated Neutralization Reveals an Achilles Heel for Norovirus.
J.Virol., 94, 2020
|
|
6XK2
 
 | |
