2IWH
 
 | | Structure of yeast Elongation Factor 3 in complex with ADPNP | | Descriptor: | ELONGATION FACTOR 3A, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION | | Authors: | Andersen, C.B.F, Becker, T, Blau, M, Anand, M, Halic, M, Balar, B, Mielke, T, Boesen, T, Pedersen, J.S, Spahn, C.M.T, Kinzy, T.G, Andersen, G.R, Beckmann, R. | | Deposit date: | 2006-06-30 | | Release date: | 2006-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Eef3 and the Mechanism of Transfer RNA Release from the E-Site.
Nature, 443, 2006
|
|
2WTD
 
 | | Crystal structure of Chk2 in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-AMINO-5-(1,3-BENZODIOXOL-4-YL)PYRIDIN-3-YL]BENZAMIDE, NITRATE ION, ... | | Authors: | Hilton, S, Naud, S, Caldwell, J.J, Boxall, K, Burns, S, Anderson, V.E, Antoni, L, Allen, C.E, Pearl, L.H, Oliver, A.W, Aherne, G.W, Garrett, M.D, Collins, I. | | Deposit date: | 2009-09-15 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification and Characterisation of 2-Aminopyridine Inhibitors of Checkpoint Kinase 2
Bioorg.Med.Chem., 18, 2010
|
|
1TQ4
 
 | | Crystal Structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, interferon-inducible GTPase | | Authors: | Ghosh, A, Uthaiah, R, Howard, J, Herrmann, C, Wolf, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of IIGP1; A Paradigm for Interferon-Inducible p47 Resistance GTPases
Mol.Cell, 15, 2004
|
|
3O36
 
 | |
6YJU
 
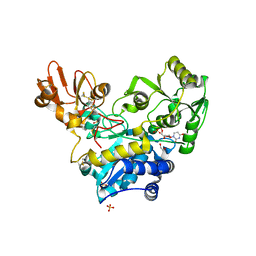 | | Crystal structure of MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with UDP and biantennary pentasaccharide M592 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
1F5U
 
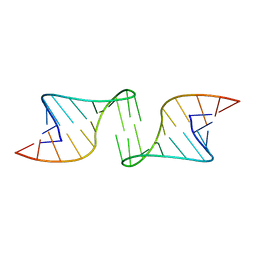 | |
1U1M
 
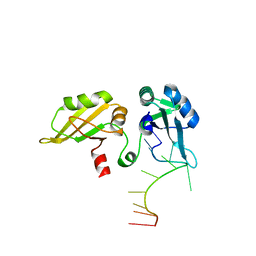 | |
1U67
 
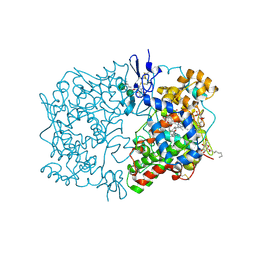 | | Crystal Structure of Arachidonic Acid Bound to a Mutant of Prostagladin H Synthase-1 that Forms Predominantly 11-HPETE. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARACHIDONIC ACID, PROTOPORPHYRIN IX CONTAINING CO, ... | | Authors: | Harman, C.A, Rieke, C.J, Garavito, R.M, Smith, W.L. | | Deposit date: | 2004-07-29 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of arachidonic Acid bound to a mutant of prostaglandin endoperoxide h synthase-1 that forms predominantly 11-hydroperoxyeicosatetraenoic Acid.
J.Biol.Chem., 279, 2004
|
|
1U1R
 
 | |
8GN9
 
 | | SPFH domain of Pyrococcus horikoshii stomatin | | Descriptor: | SODIUM ION, Stomatin homolog PH1511 | | Authors: | Komatsu, T, Matsui, I, Yokoyama, H. | | Deposit date: | 2022-08-23 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and mutational studies suggest key residues to determine whether stomatin SPFH domains form dimers or trimers.
Biochem Biophys Rep, 32, 2022
|
|
1U1L
 
 | |
1U1O
 
 | |
3W4U
 
 | | Human zeta-2 beta-2-s hemoglobin | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit beta, Hemoglobin subunit zeta, ... | | Authors: | Safo, M.K, Ko, T.-P, Russell, J.E. | | Deposit date: | 2013-01-16 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of fully liganded Hb zeta 2 beta 2(s) trapped in a tense conformation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1ZJM
 
 | | Human DNA Polymerase beta complexed with DNA containing an A-A mismatched primer terminus | | Descriptor: | 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3', 5'-D(P*GP*TP*CP*GP*G)-3', DNA polymerase beta, ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2005-04-29 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nucleotide-induced DNA polymerase active site motions accommodating a mutagenic DNA intermediate.
STRUCTURE, 13, 2005
|
|
6YH9
 
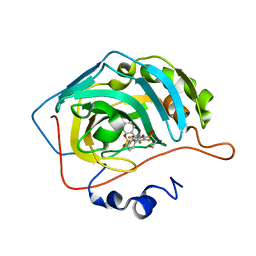 | | Crystal structure of chimeric carbonic anhydrase XII with 2,3,6-trifluoro-5-{[(1R,2S)-2-hydroxy-1,2-diphenylethyl]amino}-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 2,3,6-trifluoro-5-{[(1R,2S)-2-hydroxy-1,2-diphenylethyl]amino}-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
6YHB
 
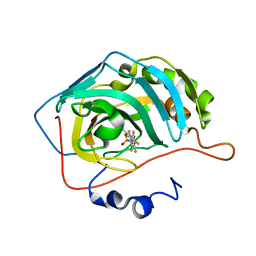 | | Crystal structure of chimeric carbonic anhydrase XII with 4-[(4,6-dimethylpyrimidin-2-yl)thio]-2,3,5,6-tetrafluorobenzenesulfonamide | | Descriptor: | 4-[(4,6-dimethylpyrimidin-2-yl)thio]-2,3,5,6-tetrafluorobenzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
6YH4
 
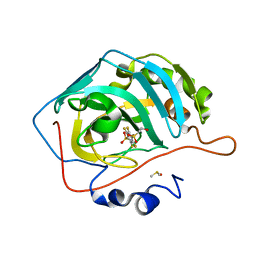 | | Crystal structure of chimeric carbonic anhydrase XII with 2,3,5,6-Tetrafluoro-4-(propylthio)benzenesulfonamide | | Descriptor: | 2,3,5,6-tetrafluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
6YH7
 
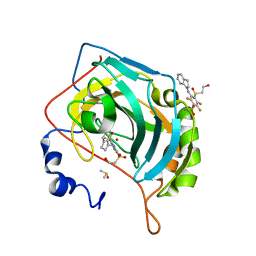 | | Crystal structure of chimeric carbonic anhydrase XII with 3-[(1S)-2,3-Dihydro-1H-inden-1-ylamino]-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 3-[(1S)-2,3-dihydro-1H-inden-1-ylamino]-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
6YH6
 
 | | Crystal structure of chimeric carbonic anhydrase XII with 2-(Cyclooctylamino)-3,5,6-trifluorobenzenesulfonamide | | Descriptor: | 2-(Cyclooctylamino)-3,5,6-trifluorobenzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
6YH5
 
 | | Crystal structure of chimeric carbonic anhydrase XII with 2-Chloro-4-[(pyrimidin-2-ylsulfanyl)acetyl]benzenesulfonamide | | Descriptor: | 2-chloro-4-[(pyrimidin-2-ylsulfanyl)acetyl]benzenesulfonamide, BENZOIC ACID, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
6YHA
 
 | | Crystal structure of chimeric carbonic anhydrase XII with 2,3,5,6-Tetrafluoro-4-(propylthio)benzenesulfonamide | | Descriptor: | 2,3,5,6-tetrafluoro-4-(propylsulfanyl)benzenesulfonamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
6YHC
 
 | | Crystal structure of chimeric carbonic anhydrase XII with 3-(benzylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 3-(benzylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
6YH8
 
 | | Crystal structure of chimeric carbonic anhydrase XII with 2-[(1S)-1,2,3,4-Tetrahydronapthalen-1-ylamino)-3,5,6-trifluorobenzenesulfonamide | | Descriptor: | 2-[(1S)-1,2,3,4-Tetrahydronapthalen-1-ylamino)-3,5,6-trifluorobenzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
8GNK
 
 | | CryoEM structure of cytosol-facing, substrate-bound ratGAT1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Nayak, S.R, Joseph, D, Penmatsa, A. | | Deposit date: | 2022-08-24 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of GABA transporter 1 reveals substrate recognition and transport mechanism.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G2F
 
 | | Crystal Structure of PRMT3 with Compound II710 | | Descriptor: | 5'-S-[3-(N'-benzylcarbamimidamido)propyl]-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-03 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A unique binding pocket induced by a noncanonical SAH mimic to develop potent and selective PRMT inhibitors.
Acta Pharm Sin B, 13, 2023
|
|
