1T86
 
 | | Crystal Structure of the Ferrous Cytochrome P450cam Mutant (L358P/C334A) | | Descriptor: | CAMPHOR, Cytochrome P450-cam, POTASSIUM ION, ... | | Authors: | Nagano, S, Tosha, T, Ishimori, K, Morishima, I, Poulos, T.L. | | Deposit date: | 2004-05-11 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the cytochrome p450cam mutant that exhibits the same spectral perturbations induced by putidaredoxin binding.
J.Biol.Chem., 279, 2004
|
|
1SWE
 
 | | APO-CORE-STREPTAVIDIN IN COMPLEX WITH BIOTIN AT PH 4.5 | | Descriptor: | BIOTIN, STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-03-04 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural studies of the streptavidin binding loop.
Protein Sci., 6, 1997
|
|
1SWD
 
 | | APO-CORE-STREPTAVIDIN IN COMPLEX WITH BIOTIN (TWO UNOCCUPIED BINDING SITES) AT PH 4.5 | | Descriptor: | BIOTIN, STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-03-04 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of the streptavidin binding loop.
Protein Sci., 6, 1997
|
|
1SWB
 
 | | APO-CORE-STREPTAVIDIN AT PH 7.5 | | Descriptor: | STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-03-04 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies of the streptavidin binding loop.
Protein Sci., 6, 1997
|
|
4TQ8
 
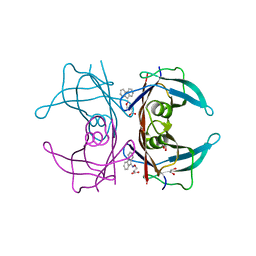 | | Dual binding mode for 3-(9H-fluoren-9-ylideneaminooxy)propanoic acid binding to Human transthyretin (TTR) | | Descriptor: | 1,2-ETHANEDIOL, 3-[(9H-fluoren-9-ylideneamino)oxy]propanoic acid, Transthyretin | | Authors: | Ciccone, L, Orlandini, E, Nencetti, S, Rossello, A, Stura, E.A. | | Deposit date: | 2014-06-10 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | X-ray crystal structure and activity of fluorenyl-based compounds as transthyretin fibrillogenesis inhibitors.
J Enzyme Inhib Med Chem, 2015
|
|
2KMB
 
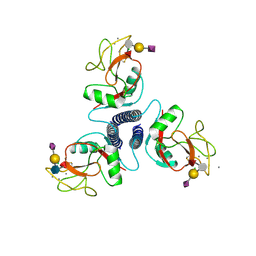 | |
1TV7
 
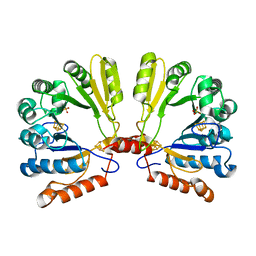 | | Structure of the S-adenosylmethionine dependent Enzyme MoaA | | Descriptor: | IRON/SULFUR CLUSTER, Molybdenum cofactor biosynthesis protein A, SULFATE ION | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2004-06-28 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
2OLA
 
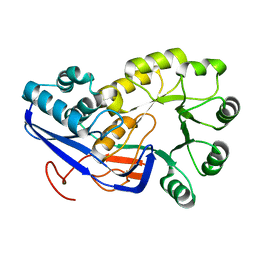 | | Crystal structure of O-succinylbenzoic acid synthetase from Staphylococcus aureus, cubic crystal form | | Descriptor: | O-succinylbenzoic acid synthetase | | Authors: | Patskovsky, Y, Sauder, J.M, Ozyurt, S, Wasserman, S.R, Smith, D, Dickey, M, Maletic, M, Reyes, C, Gheyi, T, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-18 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2ORL
 
 | | Solution structure of the cytochrome c- para-aminophenol adduct | | Descriptor: | 4-AMINOPHENOL, Cytochrome c iso-1, HEME C | | Authors: | Assfalg, M, Bertini, I, Del Conte, R, Giachetti, A, Turano, P. | | Deposit date: | 2007-02-03 | | Release date: | 2007-04-24 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Cytochrome c and organic molecules: solution structure of the p-aminophenol adduct.
Biochemistry, 46, 2007
|
|
2KKJ
 
 | |
2P85
 
 | |
8WIJ
 
 | | Crystal structure of E. coli ThrS catalytic domain mutant L489M in complex with Obafluorin | | Descriptor: | N-(2,3-dihydroxybenzoyl)-4-(4-nitrophenyl)-L-threonine, Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Wang, Z, Wang, J, Fang, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Specific glycine-dependent enzyme motion determines the potency of conformation selective inhibitors of threonyl-tRNA synthetase.
Commun Biol, 7, 2024
|
|
8WIA
 
 | | Crystal structure of E. coli ThrS catalytic domain mutant G463S | | Descriptor: | Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Wang, Z, Wang, J, Fang, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Specific glycine-dependent enzyme motion determines the potency of conformation selective inhibitors of threonyl-tRNA synthetase.
Commun Biol, 7, 2024
|
|
8WIG
 
 | | Crystal structure of E. coli ThrS catalytic domain mutant G463S/Q484A | | Descriptor: | Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Wang, Z, Wang, J, Fang, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Specific glycine-dependent enzyme motion determines the potency of conformation selective inhibitors of threonyl-tRNA synthetase.
Commun Biol, 7, 2024
|
|
6WHY
 
 | | GluN1b-GluN2B NMDA receptor in complex with GluN1 antagonist L689,560, class 1 | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-15 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
8WIH
 
 | | Crystal structure of E. coli ThrS catalytic domain mutant G463A in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Wang, Z, Wang, J, Fang, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Specific glycine-dependent enzyme motion determines the potency of conformation selective inhibitors of threonyl-tRNA synthetase.
Commun Biol, 7, 2024
|
|
4EL8
 
 | | The unliganded structure of C.bescii CelA GH48 module | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glycoside hydrolase family 48, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2012-04-10 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Revealing nature's cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA.
Science, 342, 2013
|
|
8WII
 
 | | Crystal structure of E. coli ThrS catalytic domain mutant G463A in complex with Obafluorin | | Descriptor: | Threonine--tRNA ligase, ZINC ION, ~{N}-[(2~{R},3~{S})-2-[(4-nitrophenyl)methyl]-4-oxidanylidene-oxetan-3-yl]-2,3-bis(oxidanyl)benzamide | | Authors: | Qiao, H, Wang, Z, Wang, J, Fang, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Specific glycine-dependent enzyme motion determines the potency of conformation selective inhibitors of threonyl-tRNA synthetase.
Commun Biol, 7, 2024
|
|
8ALX
 
 | | Structure of human PD-L1 in complex with inhibitor | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, AMINOMETHYLAMIDE, ACETATE ION, ... | | Authors: | Rodriguez, I, Grudnik, P, Holak, T, Magiera-Mularz, K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-16 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and biological characterization of pAC65, a macrocyclic peptide that blocks PD-L1 with equivalent potency to the FDA-approved antibodies.
Mol Cancer, 22, 2023
|
|
8ZJF
 
 | | Cryo-EM structure of human integrin alpha-E beta-7 | | Descriptor: | CALCIUM ION, Integrin alpha-E, Integrin beta-7, ... | | Authors: | Akasaka, H, Nureki, O, Kise, Y. | | Deposit date: | 2024-05-14 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure of I domain-containing integrin alpha E beta 7.
Biochem.Biophys.Res.Commun., 721, 2024
|
|
5HIW
 
 | | Sorangium cellulosum So Ce56 cytochrome P450 260B1 | | Descriptor: | Cytochrome P450 CYP260B1, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Salamanca-Pinzon, S.G, Carius, Y, Khatri, Y, Bernhardt, R, Lancaster, C.R.D. | | Deposit date: | 2016-01-12 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-function analysis for the hydroxylation of Delta 4 C21-steroids by the myxobacterial CYP260B1.
Febs Lett., 590, 2016
|
|
8WZU
 
 | |
8BAW
 
 | | X-ray structure of the CeuE Homologue from Geobacillus stearothermophilus - 5-LICAM siderophore analogue complex. | | Descriptor: | FE (III) ION, N,N'-pentane-1,5-diylbis(2,3-dihydroxybenzamide), Siderophore ABC transporter substrate-binding protein | | Authors: | Blagova, E.V, Miller, A, Booth, R, Dodson, E.J, Duhme-Klair, A.K, Wilson, K.S. | | Deposit date: | 2022-10-12 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5HRR
 
 | | HIV Integrase Catalytic Domain containing F185K + A124N + T125S mutations complexed with GSK0002 | | Descriptor: | (2S)-tert-butoxy[1-(3,4-difluorobenzyl)-6-methyl-4-(5-methyl-3,4-dihydro-2H-chromen-6-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]acetic acid, 1,2-ETHANEDIOL, CACODYLATE ION, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2016-01-24 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
6ZZW
 
 | | Structure of the N terminal domain of Bc2L-C lectin (1-131) in complex with Globo H (H-type 3) and CAS No 912569-62-1 | | Descriptor: | Lectin, SODIUM ION, [3-(2-methylimidazol-1-yl)phenyl]methanamine, ... | | Authors: | Lal, K, Bermeo, R, Imberty, A, Varrot, A. | | Deposit date: | 2020-08-05 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Prediction and Validation of a Druggable Site on Virulence Factor of Drug Resistant Burkholderia cenocepacia*.
Chemistry, 27, 2021
|
|
