5UBA
 
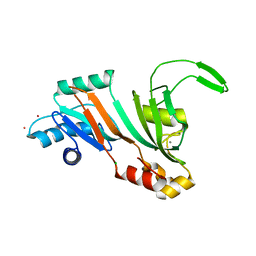 | | Human RNA Pseudouridylate Synthase Domain Containing 4 | | Descriptor: | CALCIUM ION, CHLORIDE ION, RNA pseudouridylate synthase domain-containing protein 4, ... | | Authors: | DONG, A, ZENG, H, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-12-20 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The Crystal Structure of Human Pseudouridylate Synthase Domain Containing 4. To be published
to be published
|
|
2VMY
 
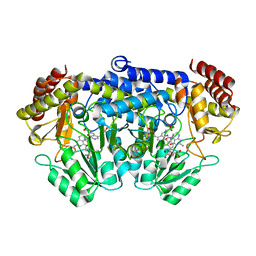 | | Crystal structure of F351GbsSHMT in complex with Gly and FTHF | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, GLYCINE, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Rajaram, V, Pai, V.R, Bisht, S, Bhavani, B.S, Appaji Rao, N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2008-01-29 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Studies of Bacillus Stearothermophilus Serine Hydroxymethyltransferase: The Role of Asn(341), Tyr(60) and Phe(351) in Tetrahydrofolate Binding.
Biochem.J., 418, 2009
|
|
1UEI
 
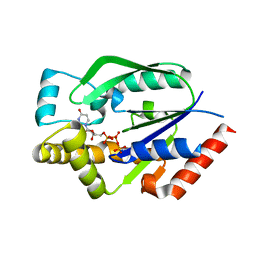 | | Crystal structure of human uridine-cytidine kinase 2 complexed with a feedback-inhibitor, UTP | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, Uridine-cytidine kinase 2 | | Authors: | Suzuki, N.N, Koizumi, K, Fukushima, M, Matsuda, A, Inagaki, F. | | Deposit date: | 2003-05-16 | | Release date: | 2004-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the specificity, catalysis, and regulation of human uridine-cytidine kinase
STRUCTURE, 12, 2004
|
|
2JYW
 
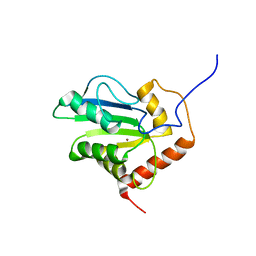 | | Solution structure of C-terminal domain of APOBEC3G | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Chen, K, Harjes, E, Gross, P.J, Fahmy, A, Lu, Y, Shindo, K, Harris, R.S, Matsuo, H. | | Deposit date: | 2007-12-20 | | Release date: | 2008-02-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G.
Nature, 452, 2008
|
|
5F2R
 
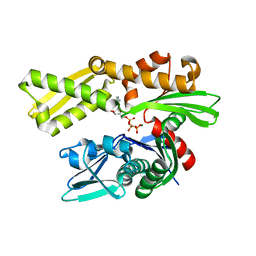 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with AMP-PCP | | Descriptor: | 78 kDa glucose-regulated protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-12-02 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5EXW
 
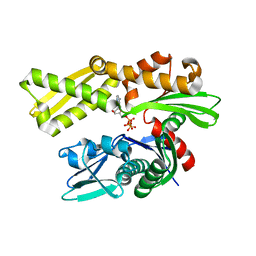 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with 7-deaza-ATP | | Descriptor: | 7-deazaadenosine-5'-triphosphate, 78 kDa glucose-regulated protein | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-24 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5F1X
 
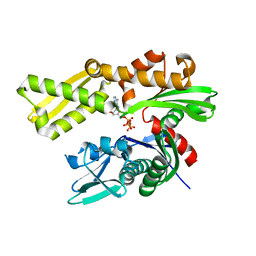 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with ATP | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-TRIPHOSPHATE | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-30 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
7AQE
 
 | | Structure of SARS-CoV-2 Main Protease bound to UNC-2327 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Meents, A. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
5EY4
 
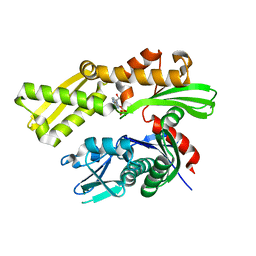 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with 2'-deoxy-ATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 78 kDa glucose-regulated protein | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-24 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
7AKQ
 
 | |
5F0X
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with 2'-deoxy-ADP and inorganic phosphate | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, 78 kDa glucose-regulated protein, ... | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-28 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
6FA3
 
 | | Antibody derived (Abd-6) small molecule binding to KRAS. | | Descriptor: | DIMETHYL SULFOXIDE, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Quevedo, C.E, Cruz-Migoni, A, Ehebauer, M.T, Carr, S.B, Phillips, S.E.V, Rabbitts, T.H. | | Deposit date: | 2017-12-15 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment.
Nat Commun, 9, 2018
|
|
6FA2
 
 | | Antibody derived (Abd-5) small molecule binding to KRAS. | | Descriptor: | 4-[2-(dimethylamino)ethoxy]-~{N}-[[(3~{R})-5-(6-methoxypyridin-2-yl)-2,3-dihydro-1,4-benzodioxin-3-yl]methyl]benzamide, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Quevedo, C.E, Cruz-Migoni, A, Ehebauer, M.T, Carr, S.B, Phillips, S.V.E, Rabbitts, T.H. | | Deposit date: | 2017-12-15 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment.
Nat Commun, 9, 2018
|
|
5EX5
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with 7-deaza-ADP and inorganic phosphate | | Descriptor: | 7-deazaadenosine-5'-diphosphate, 78 kDa glucose-regulated protein, MAGNESIUM ION, ... | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
1UCL
 
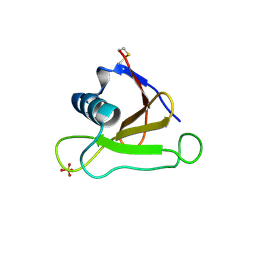 | | Mutants of RNase Sa | | Descriptor: | Guanyl-specific ribonuclease Sa, SULFATE ION | | Authors: | Takano, K, Scholtz, J.M, Sacchettini, J.C, Pace, C.N. | | Deposit date: | 2003-04-15 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The contribution of polar group burial to protein stability is strongly context-dependent
J.Biol.Chem., 278, 2003
|
|
4IDX
 
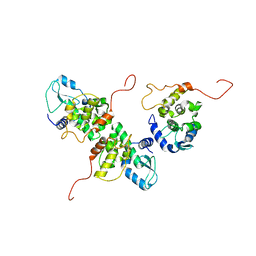 | |
7BWM
 
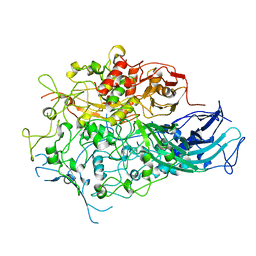 | |
6FA4
 
 | | Antibody derived (Abd-7) small molecule binding to KRAS. | | Descriptor: | 6-(2,3-dihydro-1,4-benzodioxin-5-yl)-~{N}-[4-[(dimethylamino)methyl]phenyl]-2-methoxy-pyridin-3-amine, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Quevedo, C.E, Cruz-Migoni, A, Ehebauer, M.T, Carr, S.B, Phillips, S.V.E, Rabbitts, T.H. | | Deposit date: | 2017-12-15 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment.
Nat Commun, 9, 2018
|
|
5EVZ
 
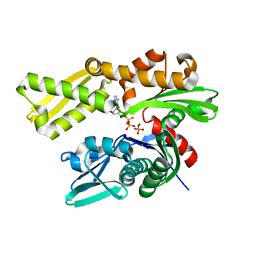 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with ADP and inorganic phosphate | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-20 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
1KDH
 
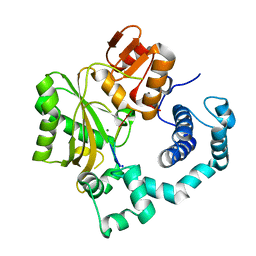 | | Binary Complex of Murine Terminal Deoxynucleotidyl Transferase with a Primer Single Stranded DNA | | Descriptor: | 5'-D(P*(BRU)P*(BRU)P*(BRU)P*(BRU))-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Delarue, M, Boule, J.B, Lescar, J, Expert-Bezancon, N, Jourdan, N, Sukumar, N, Rougeon, F, Papanicolaou, C. | | Deposit date: | 2001-11-13 | | Release date: | 2002-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase.
EMBO J., 21, 2002
|
|
6FA1
 
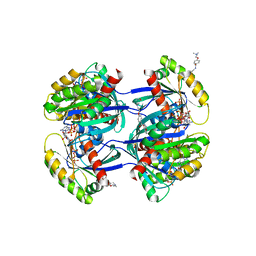 | | Antibody derived (Abd-4) small molecule binding to KRAS. | | Descriptor: | 2-[4-[[(3~{R})-2,3-dihydro-1,4-benzodioxin-3-yl]methylcarbamoyl]phenoxy]ethyl-dimethyl-azanium, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Quevedo, C.E, Cruz-Migoni, A, Ehebauer, M.T, Carr, S.B, Phillips, S.V.E, Rabbitts, T.H. | | Deposit date: | 2017-12-15 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment.
Nat Commun, 9, 2018
|
|
5YMA
 
 | | Crystal structure of ribosome assembly factor Efg1 | | Descriptor: | Putative rRNA processing protein | | Authors: | Shu, S, Ye, K. | | Deposit date: | 2017-10-21 | | Release date: | 2018-01-17 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (3.295 Å) | | Cite: | Structural and functional analysis of ribosome assembly factor Efg1.
Nucleic Acids Res., 46, 2018
|
|
3FU2
 
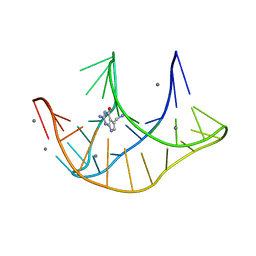 | |
1UCI
 
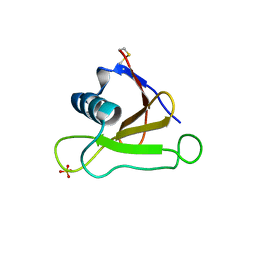 | | Mutants of RNase Sa | | Descriptor: | Guanyl-specific ribonuclease Sa, SULFATE ION | | Authors: | Takano, K, Scholtz, J.M, Sacchettini, J.C, Pace, C.N. | | Deposit date: | 2003-04-15 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The contribution of polar group burial to protein stability is strongly context-dependent
J.Biol.Chem., 278, 2003
|
|
7AIO
 
 | | Structure of Human Potassium Chloride Transporter KCC3 S45D/T940D/T997D in NaCl (Subclass) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 12 member 6, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chi, G, Man, H, Ebenhoch, R, Reggiano, G, Pike, A.C.W, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, DiMaio, F, Duerr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
