2R2Y
 
 | |
1BXS
 
 | | SHEEP LIVER CLASS 1 ALDEHYDE DEHYDROGENASE WITH NAD BOUND | | Descriptor: | ALDEHYDE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Moore, S.A, Baker, H.M, Blythe, T.J, Kitson, K.E, Kitson, T.M, Baker, E.N. | | Deposit date: | 1998-10-08 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Sheep liver cytosolic aldehyde dehydrogenase: the structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases.
Structure, 6, 1998
|
|
1BYL
 
 | |
2QQJ
 
 | |
1IFH
 
 | |
2QTY
 
 | |
2BBY
 
 | | DNA-BINDING DOMAIN FROM HUMAN RAP30, NMR, 30 STRUCTURES | | Descriptor: | RAP30 | | Authors: | Groft, C.M, Uljon, S.N, Wang, R, Werner, M.H. | | Deposit date: | 1998-04-27 | | Release date: | 1998-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural homology between the Rap30 DNA-binding domain and linker histone H5: implications for preinitiation complex assembly.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1RVW
 
 | | R STATE HUMAN HEMOGLOBIN [ALPHA V96W], CARBONMONOXY | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN, PHOSPHATE ION, ... | | Authors: | Puius, Y.A, Zou, M, Ho, N.T, Ho, C, Almo, S.C. | | Deposit date: | 1997-03-20 | | Release date: | 1998-03-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel water-mediated hydrogen bonds as the structural basis for the low oxygen affinity of the blood substitute candidate rHb(alpha 96Val-->Trp).
Biochemistry, 37, 1998
|
|
1BWY
 
 | | NMR STUDY OF BOVINE HEART FATTY ACID BINDING PROTEIN | | Descriptor: | PROTEIN (HEART FATTY ACID BINDING PROTEIN) | | Authors: | Lassen, D, Luecke, C, Kveder, M, Mesgarzadeh, A, Schmidt, J.M, Specht, B, Lezius, A, Spener, F, Rueterjans, H. | | Deposit date: | 1998-09-29 | | Release date: | 1998-10-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of bovine heart fatty-acid-binding protein with bound palmitic acid, determined by multidimensional NMR spectroscopy.
Eur.J.Biochem., 230, 1995
|
|
2GSR
 
 | |
2R5L
 
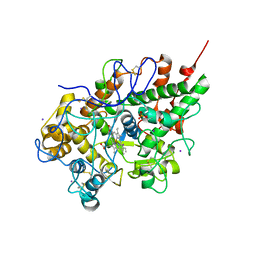 | | Crystal structure of lactoperoxidase at 2.4A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Singh, A.K, Singh, N, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Lactoperoxidase at 2.4 A Resolution.
J.Mol.Biol., 376, 2007
|
|
3UR1
 
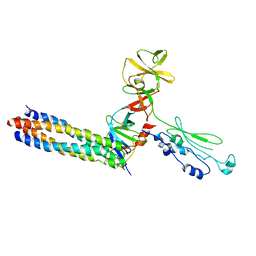 | |
4KES
 
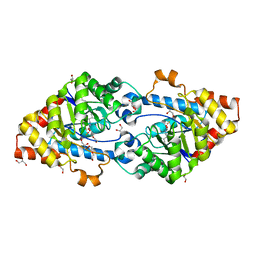 | | Crystal structure of SsoPox W263T | | Descriptor: | 1,2-ETHANEDIOL, Aryldialkylphosphatase, COBALT (II) ION, ... | | Authors: | Gotthard, G, Hiblot, J, Chabriere, E, Elias, M. | | Deposit date: | 2013-04-26 | | Release date: | 2013-10-02 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Differential Active Site Loop Conformations Mediate Promiscuous Activities in the Lactonase SsoPox.
Plos One, 8, 2013
|
|
4KFD
 
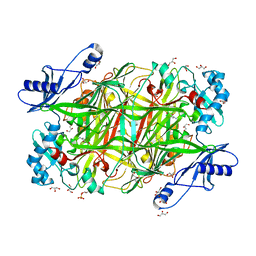 | | Crystal structure of Hansenula polymorpha copper amine oxidase-1 reduced by methylamine at pH 6.0 | | Descriptor: | COPPER (II) ION, GLYCEROL, HYDROGEN PEROXIDE, ... | | Authors: | Johnson, B.J, Yukl, E.T, Klema, V.J, Wilmot, C.M. | | Deposit date: | 2013-04-26 | | Release date: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural evidence for the semiquinone in a copper amine oxidase from Hansenula polymorpha: implications for the catalytic mechanism
J.Biol.Chem., 2013
|
|
2ORS
 
 | | Murine Inducible Nitric Oxide Synthase Oxygenase Domain (DELTA 114) 4-(Benzo[1,3]dioxol-5-yloxy)-2-(4-imidazol-1-yl-phenoxy)-6-methyl-pyrimidine Complex | | Descriptor: | 1,2-ETHANEDIOL, 4-(1,3-BENZODIOXOL-5-YLOXY)-2-[4-(1H-IMIDAZOL-1-YL)PHENOXY]-6-METHYLPYRIMIDINE, Nitric oxide synthase, ... | | Authors: | Adler, M, Whitlow, M. | | Deposit date: | 2007-02-04 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Activity of 2-Imidazol-1-ylpyrimidine Derived Inducible Nitric Oxide Synthase Dimerization Inhibitors
J.Med.Chem., 50, 2007
|
|
2BH8
 
 | | Combinatorial Protein 1b11 | | Descriptor: | 1B11 | | Authors: | De Bono, S, Riechmann, L, Girard, E, Williams, R.L, Winter, G. | | Deposit date: | 2005-01-07 | | Release date: | 2005-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Segment of Cold Shock Protein Directs the Folding of a Combinatorial Protein
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BPA
 
 | | ATOMIC STRUCTURE OF SINGLE-STRANDED DNA BACTERIOPHAGE PHIX174 AND ITS FUNCTIONAL IMPLICATIONS | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*C)-3'), PROTEIN (SUBUNIT OF BACTERIOPHAGE PHIX174) | | Authors: | McKenna, R, Xia, D, Willingmann, P, Ilag, L.L, Krishnaswamy, S, Rossmann, M.G, Olson, N.H, Baker, T.S, Incardona, N.L. | | Deposit date: | 1991-12-03 | | Release date: | 1991-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Atomic structure of single-stranded DNA bacteriophage phi X174 and its functional implications.
Nature, 355, 1992
|
|
4X6D
 
 | | CD1a ternary complex with endogenous lipids and BK6 TCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, OLEIC ACID, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-12-08 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | alpha beta T cell antigen receptor recognition of CD1a presenting self lipid ligands.
Nat.Immunol., 16, 2015
|
|
1JG3
 
 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with adenosine & VYP(ISP)HA substrate | | Descriptor: | ADENOSINE, CHLORIDE ION, SODIUM ION, ... | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2011-07-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
3M4U
 
 | |
3VM7
 
 | | Structure of an Alpha-Amylase from Malbranchea cinnamomea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Zhou, P, Hu, S.Q, Zhou, Y, Han, P, Yang, S.Q, Jiang, Z.Q. | | Deposit date: | 2011-12-09 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Novel Multifunctional alpha-Amylase from the Thermophilic Fungus Malbranchea cinnamomea: Biochemical Characterization and Three-Dimensional Structure.
Appl Biochem Biotechnol., 170, 2013
|
|
1SNM
 
 | |
2VHZ
 
 | |
1JNB
 
 | | CONNECTOR PROTEIN FROM BACTERIOPHAGE PHI29 | | Descriptor: | UPPER COLLAR PROTEIN | | Authors: | Simpson, A.A, Leiman, P.G, Tao, Y, He, Y, Badasso, M.O, Jardine, P.J, Anderson, D.L, Rossmann, M.G. | | Deposit date: | 2001-07-23 | | Release date: | 2001-08-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure determination of the head-tail connector of bacteriophage phi29.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2VHW
 
 | |
