3SUG
 
 | | Crystal structure of NS3/4A protease variant A156T in complex with MK-5172 | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-di oxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadec ino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
6CW6
 
 | | Structure of alpha-GC[8,18] bound by CD1d and in complex with the Va14Vb8.2 TCR | | Descriptor: | (2S,3S,4R)-N-OCTANOYL-1-[(ALPHA-D-GALACTOPYRANOSYL)OXY]-2-AMINO-OCTADECANE-3,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J, Zajonc, D. | | Deposit date: | 2018-03-29 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
3SV9
 
 | | Crystal structure of NS3/4A protease variant A156T in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, GLYCEROL, NS3 protease, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-12 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SVH
 
 | | Crystal Structure of the bromdomain of human CREBBP in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-ethoxybenzoic acid, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Hewings, S.D, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Conway, S.J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-07-12 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the bromdomain of human CREBBP in complex with a 3,5-dimethylisoxazol ligand
To be Published
|
|
3SVC
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: chloride complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Determination of engineered chloride-binding site structures in fluorescent proteins reveals principles of halide recognition
To be Published
|
|
3GG7
 
 | | Crystal structure of an uncharacterized metalloprotein from Deinococcus radiodurans | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, MANGANESE (II) ION, ... | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Tang, B.K, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized metalloprotein from Deinococcus radiodurans
To be Published
|
|
3SXW
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR69. | | Descriptor: | Glucose-6-phosphate isomerase | | Authors: | Vorobiev, S, Su, M, Nivon, L, Seetharaman, J, Patel, P, Xiao, R, Maglaqui, M, Ciccosanti, C, Baker, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR69.
To be Published
|
|
6CXE
 
 | | Structure of alpha-GSA[26,6P] bound by CD1d and in complex with the Va14Vb8.2 TCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Wang, J, Zajonc, D. | | Deposit date: | 2018-04-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
3SYK
 
 | | Crystal structure of the AAA+ protein CbbX, selenomethionine structure | | Descriptor: | Protein CbbX, SULFATE ION | | Authors: | Mueller-Cajar, O, Stotz, M, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2011-07-18 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase.
Nature, 479, 2011
|
|
3SW4
 
 | |
3GIB
 
 | |
3T0P
 
 | |
3T16
 
 | |
4W2I
 
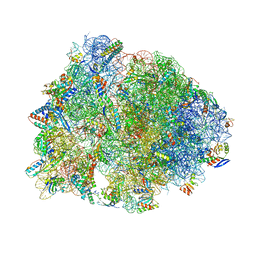 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with negamycin, mRNA and three deacylated tRNAs in the A, P and E sites | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Szal, T, Jiang, F, Gupta, P, Matsuda, R, Shiozuka, M, Steitz, T.A, Vazquez-Laslop, N, Mankin, A.S. | | Deposit date: | 2014-09-12 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Negamycin Interferes with Decoding and Translocation by Simultaneous Interaction with rRNA and tRNA.
Mol.Cell, 56, 2014
|
|
3T1D
 
 | |
3T29
 
 | | TMAO-grown trigonal trypsin (bovine) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Cahn, J, Venkat, M, Marshall, H, Juers, D. | | Deposit date: | 2011-07-22 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The use of trimethylamine N-oxide as a primary precipitating agent and related methylamine osmolytes as cryoprotective agents for macromolecular crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3T0Q
 
 | | Motor Domain Structure of the Kar3-like kinesin from Ashbya gossypii | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, AGR253Wp, ... | | Authors: | Duan, D, Hnatchuk, D.J, Brenner, J, Davis, D, Allingham, J.S. | | Deposit date: | 2011-07-20 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the Kar3-like kinesin motor domain from the filamentous fungus Ashbya gossypii.
Proteins, 80, 2012
|
|
3T11
 
 | | Dimeric inhibitor of HIV-1 protease. | | Descriptor: | (3S,11S)-11-(3-chloro-4-hydroxy-5-methoxyphenyl)-3-phenyl-2,3,4,5,10,11-hexahydro-1H-dibenzo[b,e][1,4]diazepin-1-one, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Brynda, J, Rezacova, P, Saskova, G.K, Kozisek, M, Konvalinka, J. | | Deposit date: | 2011-07-21 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Dimeric inhibitor of HIV-1 protease.
To be Published
|
|
3T2J
 
 | | Tetragonal thermolysin in the presence of betaine | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Cahn, J, Hti Lar Seng, N.S, Juers, D. | | Deposit date: | 2011-07-22 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The use of trimethylamine N-oxide as a primary precipitating agent and related methylamine osmolytes as cryoprotective agents for macromolecular crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3GGZ
 
 | |
3G3P
 
 | |
3T48
 
 | |
3GHF
 
 | | Crystal structure of the septum site-determining protein minC from Salmonella typhimurium | | Descriptor: | CITRIC ACID, Septum site-determining protein minC | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Chang, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-03 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the septum site-determining protein minC from Salmonella typhimurium
To be Published
|
|
3T4H
 
 | | Crystal Structure of AlkB in complex with Fe(III) and N-Oxalyl-S-(3-nitrobenzyl)-L-cysteine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alpha-ketoglutarate-dependent dioxygenase AlkB, FE (III) ION, ... | | Authors: | Ma, J, Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2011-07-26 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dynamic combinatorial mass spectrometry leads to inhibitors of a 2-oxoglutarate-dependent nucleic Acid demethylase.
J.Med.Chem., 55, 2012
|
|
3G16
 
 | |
