8QBY
 
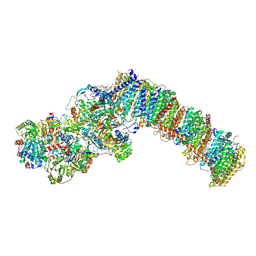 | | Respiratory complex I from Paracoccus denitrificans in MSP2N2 nanodiscs | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Ivanov, B.S, Bridges, H.R, Hirst, J. | | Deposit date: | 2023-08-25 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structure of the turnover-ready state of an ancestral respiratory complex I.
Nat Commun, 15, 2024
|
|
4EII
 
 | |
4YR7
 
 | |
1TGS
 
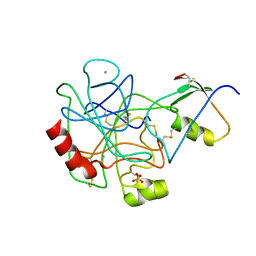 | | THREE-DIMENSIONAL STRUCTURE OF THE COMPLEX BETWEEN PANCREATIC SECRETORY INHIBITOR (KAZAL TYPE) AND TRYPSINOGEN AT 1.8 ANGSTROMS RESOLUTION. STRUCTURE SOLUTION, CRYSTALLOGRAPHIC REFINEMENT AND PRELIMINARY STRUCTURAL INTERPRETATION | | Descriptor: | CALCIUM ION, PANCREATIC SECRETORY TRYPSIN INHIBITOR (KAZAL TYPE), SULFATE ION, ... | | Authors: | Bolognesi, M, Gatti, G, Menegatti, E, Guarneri, M, Marquart, M, Papamokos, E, Huber, R. | | Deposit date: | 1982-09-27 | | Release date: | 1983-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of the complex between pancreatic secretory trypsin inhibitor (Kazal type) and trypsinogen at 1.8 A resolution. Structure solution, crystallographic refinement and preliminary structural interpretation.
J.Mol.Biol., 162, 1982
|
|
8HL7
 
 | |
1TRY
 
 | | STRUCTURE OF INHIBITED TRYPSIN FROM FUSARIUM OXYSPORUM AT 1.55 ANGSTROMS | | Descriptor: | ISOPROPYL ALCOHOL, PHOSPHORYLISOPROPANE, TRYPSIN | | Authors: | Rypniewski, W.R, Dambmann, C, Von Der Osten, C, Dauter, M, Wilson, K.S. | | Deposit date: | 1994-03-07 | | Release date: | 1996-01-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of inhibited trypsin from Fusarium oxysporum at 1.55 A.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
9D3V
 
 | | CRYSTAL STRUCTURE OF EGFR(L858R/T790M/C797S) IN COMPLEX WITH AUR-3418 | | Descriptor: | (4S)-N-[2-(4-methoxypiperidin-1-yl)pyrimidin-4-yl]-2-methyl-1-(propan-2-yl)-1H-imidazo[1,2-b]pyrazol-6-amine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Bell, J.A. | | Deposit date: | 2024-08-12 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Discovery of a Novel Mutant-Selective Epidermal Growth Factor Receptor Inhibitor Using an In Silico Enabled Drug Discovery Platform.
J.Med.Chem., 67, 2024
|
|
4Y5A
 
 | | Endothiapepsin in complex with fragment 206 | | Descriptor: | 1,2-ETHANEDIOL, 1-[5-(trifluoromethyl)furan-2-yl]methanamine, ACETATE ION, ... | | Authors: | Ehrmann, F.R, Huschmann, F.U, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallographic Fragment Sreening of an Entire Library
To Be Published
|
|
6DXZ
 
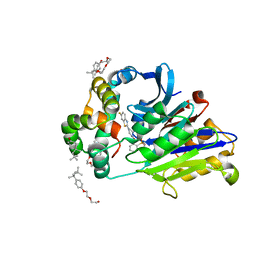 | | Rabbit N-acylethanolamine-hydrolyzing acid amidase (NAAA) in complex with non-covalent benzothiazole-piperazine inhibitor ARN19702, in presence of Triton X-100 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[4-(1,1,3,3-TETRAMETHYLBUTYL)PHENOXY]ETHOXY}ETHANOL, 4-(2,4,4-trimethylpentan-2-yl)phenol, ... | | Authors: | Gorelik, A, Gebai, A, Illes, K, Piomelli, D, Nagar, B. | | Deposit date: | 2018-07-01 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular mechanism of activation of the immunoregulatory amidase NAAA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2NAP
 
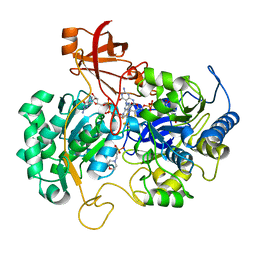 | | DISSIMILATORY NITRATE REDUCTASE (NAP) FROM DESULFOVIBRIO DESULFURICANS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Dias, J.M, Than, M, Humm, A, Huber, R, Bourenkov, G, Bartunik, H, Bursakov, S, Calvete, J, Caldeira, J, Carneiro, C, Moura, J, Moura, I, Romao, M.J. | | Deposit date: | 1998-09-18 | | Release date: | 1999-09-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the first dissimilatory nitrate reductase at 1.9 A solved by MAD methods.
Structure Fold.Des., 7, 1999
|
|
8QOC
 
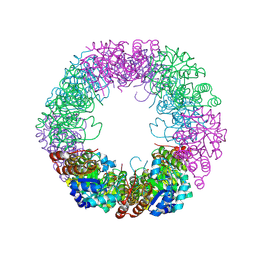 | | Crystal structure of Staphylococcus aureus PLP Synthase (Pdx1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ullah, N, Wrenger, C, Betzel, C. | | Deposit date: | 2023-09-28 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure and dynamics of the staphylococcal pyridoxal 5-phosphate synthase complex reveal transient interactions at the enzyme interface.
J.Biol.Chem., 300, 2024
|
|
1TRN
 
 | |
3H6U
 
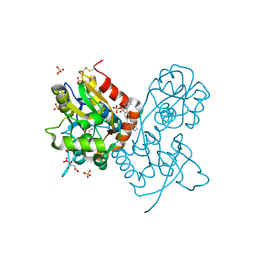 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS1493 at 1.85 A resolution | | Descriptor: | (3S)-3-cyclopentyl-6-methyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, CITRATE ANION, GLUTAMIC ACID, ... | | Authors: | Hald, H, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-04-24 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
4EUS
 
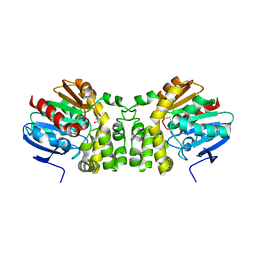 | |
4MPW
 
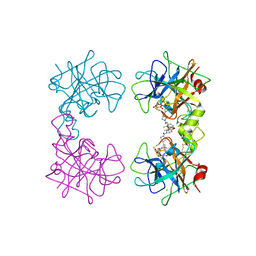 | | Human beta-tryptase co-crystal structure with [(1,1,3,3-tetramethyldisiloxane-1,3-diyl)di-1-benzofuran-3,5-diyl]bis({4-[3-(aminomethyl)phenyl]piperidin-1-yl}methanone) | | Descriptor: | ACETATE ION, CHLORIDE ION, TRIETHYLENE GLYCOL, ... | | Authors: | White, A, Stein, A.J, Suto, R. | | Deposit date: | 2013-09-13 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Novel, Nonpeptidic, Orally Active Bivalent Inhibitor of Human beta-Tryptase.
Pharmacology, 102, 2018
|
|
2N7Y
 
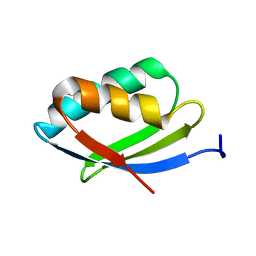 | | NMR structure of metal-binding domain 1 of ATP7B | | Descriptor: | Copper-transporting ATPase 2 | | Authors: | Yu, C, Lee, W, Dmitriev, O. | | Deposit date: | 2015-09-27 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of Metal Binding Domain 1 of the Copper Transporter ATP7B Reveals Mechanism of a Singular Wilson Disease Mutation.
Sci Rep, 8, 2018
|
|
7AK6
 
 | | Cryo-EM structure of ND6-P25L mutant respiratory complex I from Mus musculus at 3.8 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yin, Z, Bridges, H.R, Grba, D, Hirst, J. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for a complex I mutation that blocks pathological ROS production.
Nat Commun, 12, 2021
|
|
4EB8
 
 | | Crystal structure of purine nucleoside phosphorylase (W16Y, W94Y, W178Y, H257W) mutant from human complexed with DADMe-ImmG and phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, ... | | Authors: | Ho, M.C, Haapalainen, A.M, Suarez, J.J, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-03-23 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic Site Conformations in Human PNP by (19)F-NMR and Crystallography.
Chem.Biol., 20, 2013
|
|
4YRR
 
 | |
4EHM
 
 | | RabGGTase in complex with covalently bound Psoromic acid | | Descriptor: | 4-formyl-3-hydroxy-8-methoxy-1,9-dimethyl-11-oxo-11H-dibenzo[b,e][1,4]dioxepine-6-carboxylic acid, CALCIUM ION, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Deraeve, C, Bon, R.S, Alexandrov, K, Waldmann, H, Wu, Y.W, Goody, R.S, Blankenfeldt, W. | | Deposit date: | 2012-04-02 | | Release date: | 2012-05-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Psoromic Acid is a Selective and Covalent Rab-Prenylation Inhibitor Targeting Autoinhibited RabGGTase
J.Am.Chem.Soc., 134, 2012
|
|
7WWX
 
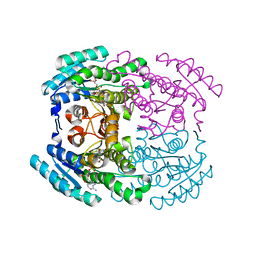 | | Crystal structure of Herbaspirillum huttiense L-arabinose 1-dehydrogenase (NAD bound form) | | Descriptor: | DI(HYDROXYETHYL)ETHER, NAD(P)-dependent dehydrogenase (Short-subunit alcohol dehydrogenase family), NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Matsubara, R, Yoshiwara, K, Watanabe, Y, Watanabe, S. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structure of L-arabinose 1-dehydrogenase as a short-chain reductase/dehydrogenase protein.
Biochem.Biophys.Res.Commun., 604, 2022
|
|
4EJ8
 
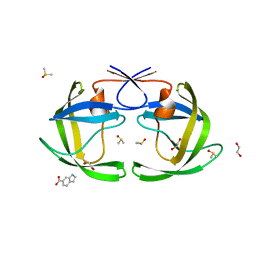 | |
9CK2
 
 | | Human PU.1 minimal ETS Domain (165-258) bound to d(AATAAGCGGAAGTGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*GP*CP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*GP*CP*TP*TP*AP*T)-3'), SODIUM ION, ... | | Authors: | Poon, G.M.K. | | Deposit date: | 2024-07-08 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Human PU.1 minimal ETS Domain (165-258) bound to d(AATAAGCGGAAGTGGG)
To Be Published
|
|
9KYG
 
 | | Crystal structure of E.coli ac4C amidohydrolase YqfB in complex with cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, MAGNESIUM ION, N(4)-acetylcytidine amidohydrolase | | Authors: | Guo, W.T, Meng, C.Y, Wen, Y, Wu, B.X. | | Deposit date: | 2024-12-09 | | Release date: | 2025-08-20 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural analysis of ASCH domain-containing proteins and their implications for nucleotide processing.
Structure, 2025
|
|
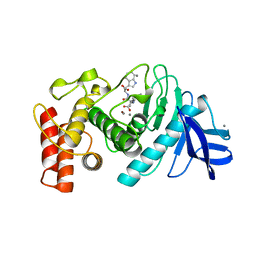
1TMN
 
 | |
