7N9J
 
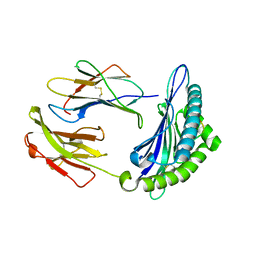 | | Crystal structure of H2DB in complex with HSF2 melanoma neoantigen | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Patskovsky, Y, Finnigan, J, Patskovska, L, Newman, J, Bhardwaj, N, Krogsgaard, M. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure of the complex between H2DB and melanoma HSF2 neoantigen YGFRNVVHI
To be Published
|
|
7NA5
 
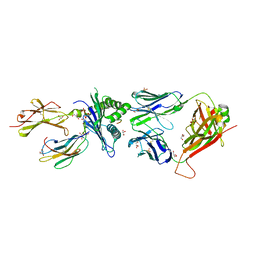 | | Structure of the H2DB-TCR ternary complex with HSF2 melanoma neoantigen | | Descriptor: | 47BE7 TCR alpha chain, 47BE7 TCR beta chain, Beta-2-microglobulin, ... | | Authors: | Patskovsky, Y, Finnigan, J, Patskovska, L, Newman, J, Bhardwaj, N, Krogsgaard, M. | | Deposit date: | 2021-06-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the TCR-H2DB ternary complex with melanoma HSF2 neoantigen YGFRNVVHI
To be Published
|
|
7N9V
 
 | |
7NAE
 
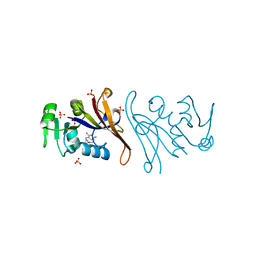 | | Crystal structure of Escherichia coli dihydrofolate reductase in complex with TRIMETHOPRIM | | Descriptor: | Dihydrofolate reductase, SULFATE ION, TRIMETHOPRIM | | Authors: | Estrada, A, Wright, D, Krucinska, J, Erlandsen, H. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
7N53
 
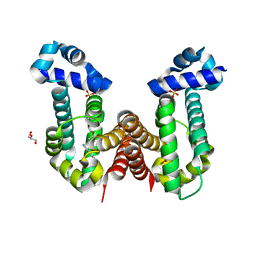 | | Complex structure of PAP4 with papaverine | | Descriptor: | 1-(3,4-DIMETHOXYBENZYL)-6,7-DIMETHOXYISOQUINOLINE, GLYCEROL, PAP4, ... | | Authors: | Kim, W, Zhang, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Using fungible biosensors to evolve improved alkaloid biosyntheses.
Nat.Chem.Biol., 18, 2022
|
|
3K94
 
 | | Crystal Structure of Thiamin pyrophosphokinase from Geobacillus thermodenitrificans, Northeast Structural Genomics Consortium Target GtR2 | | Descriptor: | Thiamin pyrophosphokinase | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Janjua, J, Xiao, R, Patel, D.J, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-15 | | Release date: | 2010-02-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Northeast Structural Genomics Consortium Target GtR2
To be Published
|
|
7N4W
 
 | | Complex structure of ROTU4 with rotundine | | Descriptor: | GLYCEROL, ROTU4, SULFATE ION, ... | | Authors: | Kim, W, Zhang, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Using fungible biosensors to evolve improved alkaloid biosyntheses.
Nat.Chem.Biol., 18, 2022
|
|
7N4Z
 
 | | Complex structure of NOS4 with noscapine | | Descriptor: | NOS4, SULFATE ION, noscapine | | Authors: | Kim, W, Zhang, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Using fungible biosensors to evolve improved alkaloid biosyntheses.
Nat.Chem.Biol., 18, 2022
|
|
7N2T
 
 | | O-acetylserine sulfhydrylase from Citrullus vulgaris in the internal aldimine state, with citrate bound | | Descriptor: | CITRIC ACID, Cysteine synthase, PENTAETHYLENE GLYCOL, ... | | Authors: | Smith, J.L, Buller, A.R, Bingman, C.A. | | Deposit date: | 2021-05-29 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Investigation of beta-Substitution Activity of O-Acetylserine Sulfhydrolase from Citrullus vulgaris.
Chembiochem, 23, 2022
|
|
7N7V
 
 | | Crystal structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes at 2 A. | | Descriptor: | CHLORIDE ION, FE (II) ION, Predicted hydroxylase | | Authors: | Han, L, Xu, W, Ma, M, Miller, M.D, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2021-06-11 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes.
To Be Published
|
|
7N54
 
 | | Complex structure of GLAU4 with glaucine | | Descriptor: | GLAU4, SULFATE ION, glaucine | | Authors: | Kim, W, Zhang, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Using fungible biosensors to evolve improved alkaloid biosyntheses.
Nat.Chem.Biol., 18, 2022
|
|
7MZQ
 
 | | Crystal structure of the UcaD lectin-binding domain in complex with fucose | | Descriptor: | CHLORIDE ION, Fimbrial adhesin UcaD, beta-L-fucopyranose | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
7MWH
 
 | | Crystal structure of BAZ2A with DNA | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, DNA (5'-D(*CP*GP*GP*AP*AP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*(5CM)P*AP*TP*TP*CP*CP*G)-3'), ... | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the BAZ2B TAM domain.
Heliyon, 8, 2022
|
|
7N91
 
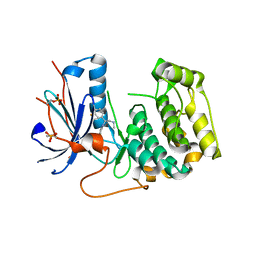 | | P70 S6K1 IN COMPLEX WITH MSC2317067A-1 | | Descriptor: | 4-{[(1S)-1-(3-fluorophenyl)-2-(methylamino)ethyl]amino}quinazoline-8-carboxamide, Ribosomal protein S6 kinase beta-1 | | Authors: | Mochalkin, I. | | Deposit date: | 2021-06-16 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of Clinical Candidate M2698, a Dual p70S6K and Akt Inhibitor, for Treatment of PAM Pathway-Altered Cancers.
J.Med.Chem., 64, 2021
|
|
7N93
 
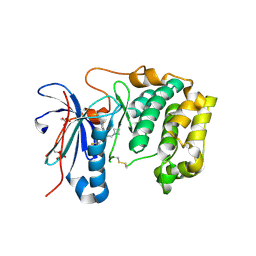 | | P70 S6K1 IN COMPLEX WITH MSC2363318A-1 | | Descriptor: | 4-({(1S)-2-(azetidin-1-yl)-1-[4-chloro-3-(trifluoromethyl)phenyl]ethyl}amino)quinazoline-8-carboxamide, Ribosomal protein S6 kinase beta-1 | | Authors: | Mochalkin, I. | | Deposit date: | 2021-06-16 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Identification of Clinical Candidate M2698, a Dual p70S6K and Akt Inhibitor, for Treatment of PAM Pathway-Altered Cancers.
J.Med.Chem., 64, 2021
|
|
7N9O
 
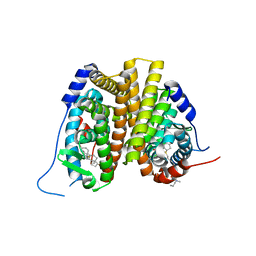 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with Aliphatic SERD S-C10(15) | | Descriptor: | 10-{[3,17beta-dihydroxyestra-1,3,5(10)-trien-7beta-yl]sulfanyl}-N-methyl-N-propyldecanamide, Estrogen receptor | | Authors: | Diennet, M, El Ezzy, M, Thiombane, K, Cotnoir-White, D, Poupart, J, Gao, Z, Mendoza, S.R, Marinier, A, Gleason, J, Mader, S.C, Fanning, S.W. | | Deposit date: | 2021-06-18 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Estrogen Receptor Alpha Ligand Binding Domain in Complex with Aliphatic SERD S-C10(15)
To Be Published
|
|
7MZS
 
 | | Crystal structure of the UcaD lectin-binding domain in complex with galactose | | Descriptor: | CHLORIDE ION, Fimbrial adhesin UcaD, alpha-D-galactopyranose | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
3JZA
 
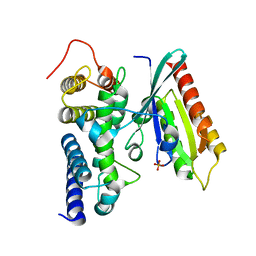 | | Crystal structure of human Rab1b in complex with the GEF domain of DrrA/SidM from Legionella pneumophila | | Descriptor: | PHOSPHATE ION, Ras-related protein Rab-1B, Uncharacterized protein DrrA | | Authors: | Schoebel, S, Oesterlin, L.K, Blankenfeldt, W, Goody, R.S, Itzen, A. | | Deposit date: | 2009-09-23 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity.
Mol.Cell, 36, 2009
|
|
7MZR
 
 | | Crystal structure of the UcaD lectin-binding domain in complex with glucose | | Descriptor: | CHLORIDE ION, Fimbrial adhesin UcaD, beta-D-glucopyranose | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
7N1M
 
 | | Crystal Structure of the Class D Beta-lactamase OXA-935 from Pseudomonas aeruginosa, Orthorhombic Crystal Form | | Descriptor: | Beta-lactamase OXA-935, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.B, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-27 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Functional and Structural Characterization of OXA-935, a Novel OXA-10-Family beta-Lactamase from Pseudomonas aeruginosa.
Antimicrob.Agents Chemother., 66, 2022
|
|
7MST
 
 | | Phosphorylated human E105Qa GTP-specific succinyl-CoA synthetase complexed with coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, GLYCEROL, ... | | Authors: | Huang, J, Fraser, M.E. | | Deposit date: | 2021-05-12 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The structure of succinyl-CoA synthetase bound to the succinyl-phosphate intermediate clarifies the catalytic mechanism of ATP-citrate lyase
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7N5Q
 
 | |
7N7G
 
 | | Crystal Structure of FosB from Enterococcus faecium with Fosfomycin | | Descriptor: | 1,2-ETHANEDIOL, FOSFOMYCIN, MANGANESE (II) ION, ... | | Authors: | Shay, M.R, Simmons, Z, Thompson, M.K. | | Deposit date: | 2021-06-10 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional characterization of fosfomycin resistance conferred by FosB from Enterococcus faecium.
Protein Sci., 31, 2022
|
|
3K71
 
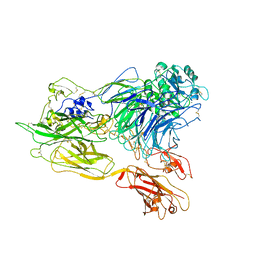 | | Structure of integrin alphaX beta2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xie, C, Zhu, J, Chen, X, Mi, L, Nishida, N, Springer, T.A. | | Deposit date: | 2009-10-11 | | Release date: | 2010-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structure of an integrin with an alphaI domain, complement receptor type 4.
Embo J., 29, 2010
|
|
7MR9
 
 | |
