1RH5
 
 | | The structure of a protein conducting channel | | Descriptor: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, SecBeta | | Authors: | van den Berg, B, Clemons Jr, W.M, Collinson, I, Modis, Y, Hartmann, E, Harrison, S.C, Rapoport, T.A. | | Deposit date: | 2003-11-13 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structure of a protein-conducting channel
Nature, 427, 2004
|
|
1RH6
 
 | | Bacteriophage Lambda Excisionase (Xis)-DNA Complex | | Descriptor: | 5'-D(*CP*TP*AP*TP*GP*TP*AP*GP*TP*CP*TP*GP*TP*TP*G)-3', 5'-D(P*CP*AP*AP*CP*AP*GP*AP*CP*TP*AP*CP*AP*TP*AP*G)-3', Excisionase | | Authors: | Sam, M.D, Cascio, D, Johnson, R.C, Clubb, R.T. | | Deposit date: | 2003-11-13 | | Release date: | 2004-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the excisionase-DNA complex from bacteriophage lambda.
J.Mol.Biol., 338, 2004
|
|
1RH7
 
 | | Crystal Structure of Resistin-like beta | | Descriptor: | HEXAETHYLENE GLYCOL, PLATINUM (II) ION, Resistin-like beta | | Authors: | Patel, S.D, Rajala, M.W, Scherer, P.E, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-13 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Disulfide-dependent multimeric assembly of resistin family hormones
Science, 304, 2004
|
|
1RH8
 
 | | Three-dimensional structure of the calcium-free Piccolo C2A-domain | | Descriptor: | Piccolo protein | | Authors: | Garcia, J, Gerber, S.H, Sugita, S, Sudhof, T.C, Rizo, J. | | Deposit date: | 2003-11-14 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A conformational switch in the Piccolo C2A domain regulated by alternative splicing.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1RH9
 
 | | Family GH5 endo-beta-mannanase from Lycopersicon esculentum (tomato) | | Descriptor: | endo-beta-mannanase | | Authors: | Oakley, A.J, Bourgault, R, Bewley, J.D, Wilce, M.C.J. | | Deposit date: | 2003-11-14 | | Release date: | 2005-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional structure of (1,4)-beta-D-mannan mannanohydrolase from tomato fruit.
Protein Sci., 14, 2005
|
|
1RHA
 
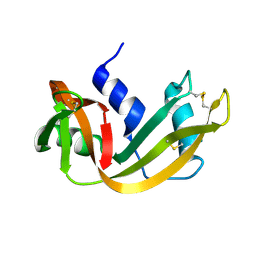 | | WATER DEPENDENT DOMAIN MOTION AND FLEXIBILITY IN RIBONUCLEASE A AND THE INVARIANT FEATURES IN ITS HYDRATION SHELL. AN X-RAY STUDY OF TWO LOW HUMIDITY CRYSTAL FORMS OF THE ENZYME | | Descriptor: | RIBONUCLEASE A | | Authors: | Radha Kishan, K.V, Chandra, N.R, Sudarsanakumar, C, Suguna, K, Vijayan, M. | | Deposit date: | 1994-11-13 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Water-dependent domain motion and flexibility in ribonuclease A and the invariant features in its hydration shell. An X-ray study of two low-humidity crystal forms of the enzyme.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1RHB
 
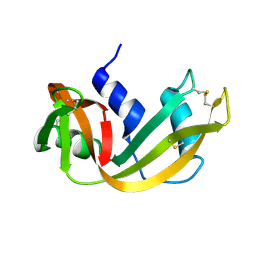 | | WATER DEPENDENT DOMAIN MOTION AND FLEXIBILITY IN RIBONUCLEASE A AND THE INVARIANT FEATURES IN ITS HYDRATION SHELL. AN X-RAY STUDY OF TWO LOW HUMIDITY CRYSTAL FORMS OF THE ENZYME | | Descriptor: | RIBONUCLEASE A | | Authors: | Radha Kishan, K.V, Chandra, N.R, Sudarsanakumar, C, Suguna, K, Vijayan, M. | | Deposit date: | 1994-11-13 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Water-dependent domain motion and flexibility in ribonuclease A and the invariant features in its hydration shell. An X-ray study of two low-humidity crystal forms of the enzyme.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1RHC
 
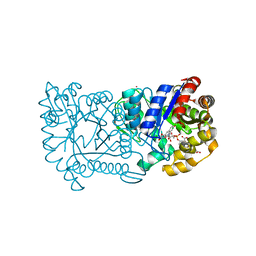 | | F420-dependent secondary alcohol dehydrogenase in complex with an F420-acetone adduct | | Descriptor: | ACETONE, CHLORIDE ION, COENZYME F420, ... | | Authors: | Aufhammer, S.W, Warkentin, E, Berk, H, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2003-11-14 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Coenzyme binding in f(420)-dependent secondary alcohol dehydrogenase, a member of the bacterial luciferase family.
Structure, 12, 2004
|
|
1RHD
 
 | |
1RHF
 
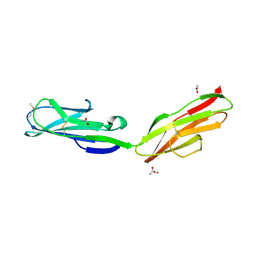 | | Crystal Structure of human Tyro3-D1D2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, Tyrosine-protein kinase receptor TYRO3, ... | | Authors: | Heiring, C, Dahlback, B, Muller, Y.A. | | Deposit date: | 2003-11-14 | | Release date: | 2004-03-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Ligand recognition and homophilic interactions in Tyro3: structural insights into the Axl/Tyro3 receptor tyrosine kinase family.
J.Biol.Chem., 279, 2004
|
|
1RHG
 
 | |
1RHH
 
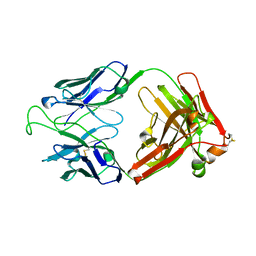 | | Crystal Structure of the Broadly HIV-1 Neutralizing Fab X5 at 1.90 Angstrom Resolution | | Descriptor: | Fab X5, heavy chain, light chain | | Authors: | Darbha, R, Phogat, S, Labrijn, A.F, Shu, Y, Gu, Y, Andrykovitch, M, Zhang, M.Y, Pantophlet, R, Martin, L, Vita, C, Burton, D.R, Dimitrov, D.S, Ji, X. | | Deposit date: | 2003-11-14 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Broadly Cross-Reactive HIV-1-Neutralizing Fab X5 and Fine Mapping of Its Epitope
Biochemistry, 43, 2004
|
|
1RHI
 
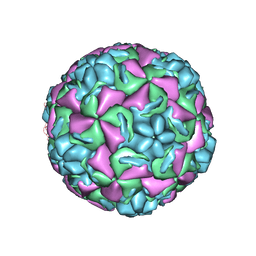 | | HUMAN RHINOVIRUS 3 COAT PROTEIN | | Descriptor: | CALCIUM ION, HUMAN RHINOVIRUS 3 COAT PROTEIN | | Authors: | Zhao, R, Rossmann, M.G. | | Deposit date: | 1996-06-17 | | Release date: | 1997-03-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human rhinovirus 3 at 3.0 A resolution.
Structure, 4, 1996
|
|
1RHJ
 
 | |
1RHK
 
 | |
1RHL
 
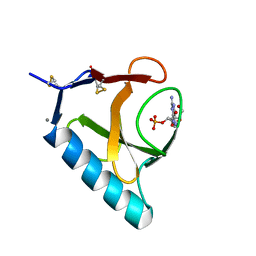 | | RIBONUCLEASE T1 COMPLEXED WITH 2'GMP/G23A MUTANT | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Huyghues-Despointes, B.M.P, Langhorst, U, Steyaert, J, Pace, C.N, Scholtz, J.M. | | Deposit date: | 1998-10-09 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Hydrogen-exchange stabilities of RNase T1 and variants with buried and solvent-exposed Ala --> Gly mutations in the helix.
Biochemistry, 38, 1999
|
|
1RHM
 
 | |
1RHO
 
 | | STRUCTURE OF RHO GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR | | Descriptor: | RHO GDP-DISSOCIATION INHIBITOR 1, SULFATE ION | | Authors: | Keep, N.H, Moody, P.C.E, Roberts, G.C.K. | | Deposit date: | 1996-10-12 | | Release date: | 1997-10-15 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A modulator of rho family G proteins, rhoGDI, binds these G proteins via an immunoglobulin-like domain and a flexible N-terminal arm.
Structure, 5, 1997
|
|
1RHP
 
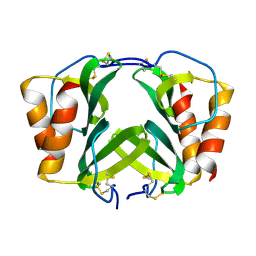 | |
1RHQ
 
 | |
1RHR
 
 | |
1RHS
 
 | | SULFUR-SUBSTITUTED RHODANESE | | Descriptor: | SULFUR-SUBSTITUTED RHODANESE | | Authors: | Zanotti, G, Gliubich, F, Colapietro, M, Barba, L. | | Deposit date: | 1997-07-16 | | Release date: | 1998-01-21 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure of sulfur-substituted rhodanese at 1.36 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1RHT
 
 | | 24-MER RNA HAIRPIN COAT PROTEIN BINDING SITE FOR BACTERIOPHAGE R17 (NMR, MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | RNA (5'-R(P*GP*GP*GP*AP*CP*UP*GP*AP*CP*GP*AP*UP*CP*AP*CP*GP*CP*AP*GP*UP*CP*UP*AP*U)-3') | | Authors: | Borer, P.N, Lin, Y, Wang, S, Roggenbuck, M.W, Gott, J.M, Uhlenbeck, O.C, Pelczer, I. | | Deposit date: | 1995-03-03 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Proton NMR and structural features of a 24-nucleotide RNA hairpin.
Biochemistry, 34, 1995
|
|
1RHU
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF CASPASE-3 WITH A 5,6,7 TRICYCLIC PEPTIDOMIMETIC INHIBITOR | | Descriptor: | (3S)-3-[({(2S)-5-[(N-ACETYL-L-ALPHA-ASPARTYL)AMINO]-4-OXO-1,2,4,5,6,7-HEXAHYDROAZEPINO[3,2,1-HI]INDOL-2-YL}CARBONYL)AMINO]-5-(BENZYLSULFANYL)-4-OXOPENTANOIC ACID, Caspase-3 | | Authors: | Becker, J.W, Rotonda, J, Soisson, S.M. | | Deposit date: | 2003-11-14 | | Release date: | 2004-05-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Reducing the Peptidyl Features of Caspase-3 Inhibitors: A Structural Analysis.
J.Med.Chem., 47, 2004
|
|
1RHW
 
 | | The solution structure of the pH-induced monomer of dynein light chain LC8 from Drosophila | | Descriptor: | Dynein light chain 1, cytoplasmic | | Authors: | Makokha, M, Huang, Y.J, Montelione, G, Edison, A.S, Barbar, E. | | Deposit date: | 2003-11-14 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the pH-induced monomer of dynein light-chain LC8 from Drosophila.
Protein Sci., 13, 2004
|
|
