4BK9
 
 | |
4BDA
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 3,4-dihydro[1,2,4]triazolo[1,5-a][3,1]benzimidazol-9-ium, CHLORIDE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BDK
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, CHECKPOINT KINASE 2, N-[(4-methoxyphenyl)methyl]quinoxaline-6-carboxamide, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
8RH0
 
 | | Trimeric HSV-1F gB ectodomain in postfusion conformation with three bound HDIT102 Fab molecules. | | Descriptor: | Envelope glycoprotein B, HDIT102 Fab heavy chain | | Authors: | Kalbermatter, D, Seyfizadeh, N, Imhof, T, Ries, M, Mueller, C, Jenner, L, Blumenschein, E, Yendrzheyevskiy, A, Moog, K, Eckert, D, Engel, R, Diebolder, P, Chami, M, Krauss, J, Schaller, T, Arndt, M. | | Deposit date: | 2023-12-14 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus.
J.Biomed.Sci., 31, 2024
|
|
8RT8
 
 | |
8RT4
 
 | |
4BI5
 
 | | CRYSTAL STRUCTURE OF A DOUBLE MUTANT (C202A AND C222D) OF TRIOSEPHOSPHATE ISOMERASE FROM GIARDIA LAMBLIA. | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Torres-Larios, A, Enriquez-Flores, S, Reyes-Vivas, H, Oria-Hernandez, J, Hernandez-Alcantara, G. | | Deposit date: | 2013-04-09 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Perturbation of Giardia Lamblia Triosephosphate Isomerase by Modification of a Non-Catalytic, Non-Conserved Region.
Plos One, 8, 2013
|
|
4BKR
 
 | | Enoyl-ACP reductase from Yersinia pestis (wildtype, removed Histag) with cofactor NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Hirschbeck, M.W, Neckles, C, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-04-29 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia Pestis Fabv Enoyl-Acp Reductase.
Biochemistry, 55, 2016
|
|
4BKG
 
 | | crystal structure of human diSUMO-2 | | Descriptor: | SMALL UBIQUITIN-RELATED MODIFIER 2 | | Authors: | Keusekotten, K, Bade, V.N, Meyer-Teschendorf, K, Sriramachandran, A, Fischer-Schrader, K, Krause, A, Horst, C, Hofmann, K, Dohmen, R.J, Praefcke, G.J.K. | | Deposit date: | 2013-04-25 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Multivalent Interactions of the Sumo-Interaction Motifs in the Ring-Finger Protein 4 (Rnf4) Determine the Specificity for Chains of the Small Ubiquitin-Related Modifier (Sumo).
Biochem.J., 457, 2014
|
|
4ARM
 
 | | Structure of the inactive pesticin T201A mutant | | Descriptor: | PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4AVW
 
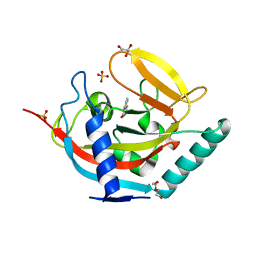 | | Crystal structure of human tankyrase 2 in complex with TIQ-A | | Descriptor: | 4H-thieno[2,3-c]isoquinolin-5-one, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2012-05-29 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Evaluation and Structural Basis for the Inhibition of Tankyrases by Parp Inhibitors
Acs Med.Chem.Lett., 5, 2014
|
|
8RZH
 
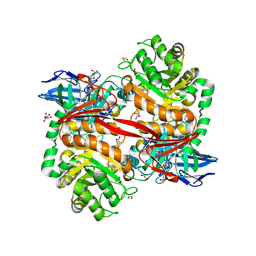 | | ZgGH129 from Zobellia galactanivorans in complex with the inhibitor AD-DGJ (3,6-anhydro-D-1-deoxygalactonojirimycin). | | Descriptor: | (1~{R},4~{S},5~{R},8~{S})-6-oxa-2-azabicyclo[3.2.1]octane-4,8-diol, 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 2024
|
|
4BIU
 
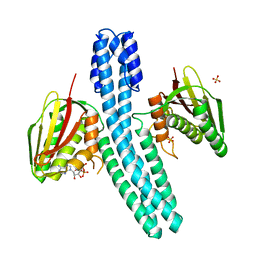 | | Crystal structure of CpxAHDC (orthorhombic form 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SENSOR PROTEIN CPXA, SULFATE ION | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-04-13 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
4BIZ
 
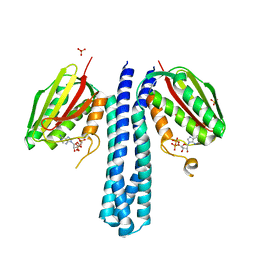 | |
4B9K
 
 | | pVHL-ELOB-ELOC complex_(2S,4R)-1-(3-amino-2-methylbenzoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide bound | | Descriptor: | (2S,4R)-1-(3-amino-2-methylbenzoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide, ACETATE ION, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 1, ... | | Authors: | Buckley, D.L, Gustafson, J.L, VanMolle, I, Roth, A.G, SeopTae, H, Gareiss, P.C, Jorgensen, W.L, Ciulli, A, Crews, C.M. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small-Molecule Inhibitors of the Interaction between the E3 Ligase Vhl and Hif1 Alpha
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4BK8
 
 | | Superoxide reductase (Neelaredoxin) from Ignicoccus hospitalis | | Descriptor: | DESULFOFERRODOXIN, FERROUS IRON-BINDING REGION, FE (III) ION | | Authors: | Romao, C.V, Matias, P.M, Pinho, F.G, Sousa, C.M, Barradas, A.R, Pinto, A.F, Teixeira, M, Bandeiras, T.M. | | Deposit date: | 2013-04-22 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Structure of a Natural Sor Mutant
To be Published
|
|
4BJS
 
 | | Crystal structure of the Rif1 C-terminal domain (Rif1-CTD) from Saccharomyces cerevisiae | | Descriptor: | TELOMERE LENGTH REGULATOR PROTEIN RIF1 | | Authors: | Bunker, R.D, Shi, T, Gut, H, Scrima, A, Thoma, N.H. | | Deposit date: | 2013-04-19 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
8RZJ
 
 | | ZgGH129 from Zobellia galactanivorans in complex with the inhibitor ADG-IF (3,6-anhydro-D-galacto-isofagomine). | | Descriptor: | (1~{R},5~{R},8~{S})-6-oxa-3-azabicyclo[3.2.1]octan-8-ol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 2024
|
|
4BBQ
 
 | | Crystal structure of the CXXC and PHD domain of Human Lysine-specific Demethylase 2A (KDM2A)(FBXL11) | | Descriptor: | 1,2-ETHANEDIOL, LYSINE-SPECIFIC DEMETHYLASE 2A, ZINC ION | | Authors: | Allerston, C.K, Watson, A.A, Edlich, C, Li, B, Chen, Y, Ball, L, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Laue, E.D, Gileadi, O. | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of the Cxxc and Phd Domain of Human Lysine-Specific Demethylase 2A (Kdm2A)(Fbxl11)
To be Published
|
|
8RTC
 
 | |
8RZI
 
 | | ZgGH129 from Zobellia galactanivorans soaked with 1,2-diF-ADG (3,6-Anhydro-2-deoxy-2-fluoro-a-D-galactopyranosyl fluoride) resulting in a trapped glycosyl-enzyme intermediate. | | Descriptor: | (1~{R},4~{S},5~{S},8~{S})-4-fluoranyl-2,6-dioxabicyclo[3.2.1]octan-8-ol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 2024
|
|
4BG7
 
 | |
4BJN
 
 | | Crystal structure of the flax-rust effector AvrM-A | | Descriptor: | AVRM-A | | Authors: | Ve, T, Williams, S.J, Kobe, B. | | Deposit date: | 2013-04-19 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the Flax-Rust Effector Avrm Reveal Insights Into the Molecular Basis of Plant-Cell Entry and Effector-Triggered Immunity
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8RZG
 
 | | ZgGH129 from Zobellia galactanivorans soaked with the product of the reaction ADG (3,6-anhydro-D-galactose). | | Descriptor: | (1~{R},4~{S},5~{R},8~{S})-2,6-dioxabicyclo[3.2.1]octane-4,8-diol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 2024
|
|
4BAX
 
 | |
