1DNF
 
 | | EFFECTS OF 5-FLUOROURACIL/GUANINE WOBBLE BASE PAIRS IN Z-DNA. MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGFG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(UFP)P*G)-3'), MAGNESIUM ION | | Authors: | Coll, M, Saal, D, Frederick, C.A, Aymami, J, Rich, A, Wang, A.H.-J. | | Deposit date: | 1988-12-12 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effects of 5-fluorouracil/guanine wobble base pairs in Z-DNA: molecular and crystal structure of d(CGCGFG).
Nucleic Acids Res., 17, 1989
|
|
6K5L
 
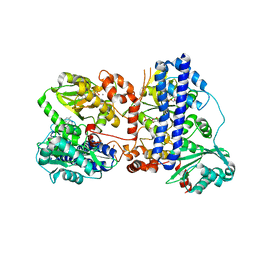 | | The crystal structure of isocitrate dehydrogenase kinase/phosphatase wtih two Mn2+ from E. coli | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Isocitrate dehydrogenase kinase/phosphatase, ... | | Authors: | Zhang, X, Lei, Z, Zheng, J, Jia, Z. | | Deposit date: | 2019-05-29 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Characterization of metal binding of bifunctional kinase/phosphatase AceK and implication in activity modulation.
Sci Rep, 9, 2019
|
|
6KPQ
 
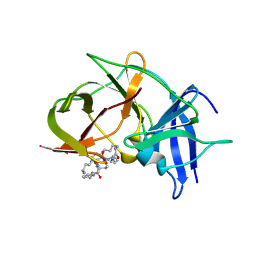 | | Crystal structure of Zika NS2B-NS3 protease with compound 8 | | Descriptor: | 1-[(10~{R},17~{S},20~{S})-17,20-bis(4-azanylbutyl)-4,9,16,19,22-pentakis(oxidanylidene)-3,8,15,18,21-pentazabicyclo[22.3.1]octacosa-1(27),24(28),25-trien-10-yl]guanidine, NS3 protease, Serine protease subunit NS2B | | Authors: | Quek, J.P. | | Deposit date: | 2019-08-15 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
3NXX
 
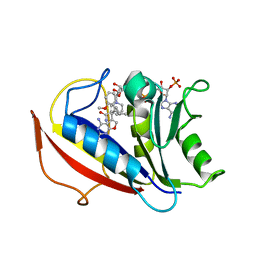 | | Preferential Selection of Isomer Binding from Chiral Mixtures: Alternate Binding Modes Observed for the E- and Z-isomers of a Series of 5-Substituted 2,4-Diaminofuro-2,3-d]pyrimidines as Ternary Complexes with NADPH and Human Dihydrofolate Reductase | | Descriptor: | 5-[(1E)-2-(2-methoxyphenyl)-4-methylpent-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V. | | Deposit date: | 2010-07-14 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Preferential selection of isomer binding from chiral mixtures: alternate binding modes observed for the E and Z isomers of a series of 5-substituted 2,4-diaminofuro[2,3-d]pyrimidines as ternary complexes with NADPH and human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3OOH
 
 | | Crystal structure of E. Coli purine nucleoside phosphorylase with PO4 | | Descriptor: | PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | Authors: | Mikleusevic, G, Stefanic, Z, Narzyk, M, Wielgus-Kutrowska, B, Bzowska, A, Luic, M. | | Deposit date: | 2010-08-31 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Validation of the catalytic mechanism of Escherichia coli purine nucleoside phosphorylase by structural and kinetic studies.
Biochimie, 93, 2011
|
|
6CCH
 
 | | NMR data-driven model of GTPase KRas-GMPPNP tethered to a nanodisc (E3 state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
4IJG
 
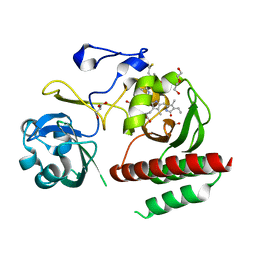 | | Crystal structure of monomeric bacteriophytochrome | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Auldridge, M.E. | | Deposit date: | 2012-12-21 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Origins of fluorescence in evolved bacteriophytochromes.
J.Biol.Chem., 289, 2014
|
|
6PQP
 
 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the covalent agonist BITC | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-benzylthioformamide, ... | | Authors: | Suo, Y, Wang, Z, Zubcevic, L, Hsu, A.L, He, Q, Borgnia, M.J, Ji, R.-R, Lee, S.-Y. | | Deposit date: | 2019-07-09 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural Insights into Electrophile Irritant Sensing by the Human TRPA1 Channel.
Neuron, 105, 2020
|
|
1WOF
 
 | | Crystal Structure Of SARS-CoV Mpro in Complex with an Inhibitor N1 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]-L-ALANYL-L-VALYL-N~1~-((1S)-4-ETHOXY-4-OXO-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Yang, H, Bartlam, M, Xue, X, Yang, K, Liang, W, Rao, Z. | | Deposit date: | 2004-08-18 | | Release date: | 2005-08-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases.
Plos Biol., 3, 2005
|
|
6PQO
 
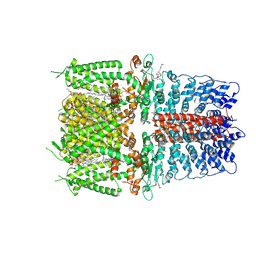 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the covalent agonist JT010 | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloro-N-[4-(4-methoxyphenyl)-1,3-thiazol-2-yl]-N-(3-methoxypropyl)acetamide, ... | | Authors: | Suo, Y, Wang, Z, Zubcevic, L, Hsu, A.L, He, Q, Borgnia, M.J, Ji, R.-R, Lee, S.-Y. | | Deposit date: | 2019-07-09 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural Insights into Electrophile Irritant Sensing by the Human TRPA1 Channel.
Neuron, 105, 2020
|
|
4INX
 
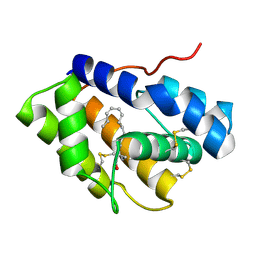 | | Structure of Pheromone-binding protein 1 in complex with (Z,Z)-11,13- hexadecadienol | | Descriptor: | (11Z,13Z)-hexadeca-11,13-dien-1-ol, Pheromone-binding protein 1 | | Authors: | di Luccio, E, Wilson, D.K. | | Deposit date: | 2013-01-07 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Crystallographic Observation of pH-Induced Conformational Changes in the Amyelois transitella Pheromone-Binding Protein AtraPBP1.
Plos One, 8, 2013
|
|
1R78
 
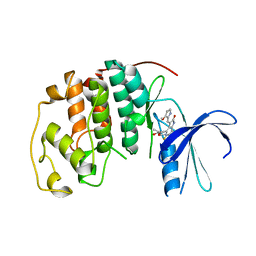 | | CDK2 complex with a 4-alkynyl oxindole inhibitor | | Descriptor: | 4-((3R,4S,5R)-4-AMINO-3,5-DIHYDROXY-HEX-1-YNYL)-5-FLUORO-3-[1-(3-METHOXY-1H-PYRROL-2-YL)-METH-(Z)-YLIDENE]-1,3-DIHYDRO-INDOL-2-ONE, Cell division protein kinase 2 | | Authors: | Luk, K.-C, Simcox, M.E, Schutt, A, Rowan, K, Thompson, T, Chen, Y, Kammlott, U, DePinto, W, Dunten, P, Dermatakis, A. | | Deposit date: | 2003-10-20 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new series of potent oxindole inhibitors of CDK2
Bioorg.Med.Chem.Lett., 14, 2004
|
|
7WLD
 
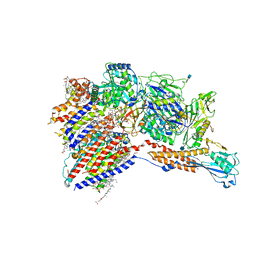 | | Cryo-EM structure of the human glycosylphosphatidylinositol transamidase complex at 2.53 Angstrom resolution | | Descriptor: | (4S,7R)-7-[(hexadecanoyloxy)methyl]-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-3,5,8-trioxa-4lambda~5~-phosphahexacosan-1-aminium, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, Y, Li, T, Luo, Y, Chao, Y, Jia, G, Zhou, Z, Su, Z, Qu, Q, Li, D. | | Deposit date: | 2022-01-13 | | Release date: | 2022-04-27 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Molecular insights into biogenesis of glycosylphosphatidylinositol anchor proteins.
Nat Commun, 13, 2022
|
|
7LUR
 
 | |
2G3J
 
 | | Structure of S.olivaceoviridis xylanase Q88A/R275A mutant | | Descriptor: | PHOSPHATE ION, Xylanase, alpha-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Diertavitian, S, Kaneko, S, Fujimoto, Z, Kuno, A, Johansson, E, Lo Leggio, L. | | Deposit date: | 2006-02-20 | | Release date: | 2007-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based engineering of glucose specificity in a family 10 xylanase from Streptomyces olivaceoviridis E-86
PROCESS BIOCHEM, 47, 2012
|
|
2G3I
 
 | | Structure of S.olivaceoviridis xylanase Q88A/R275A mutant | | Descriptor: | PHOSPHATE ION, Xylanase | | Authors: | Diertavitian, S, Kaneko, S, Fujimoto, Z, Kuno, A, Johansson, E, Lo Leggio, L. | | Deposit date: | 2006-02-20 | | Release date: | 2007-03-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based engineering of glucose specificity in a family 10 xylanase from Streptomyces olivaceoviridis E-86
PROCESS BIOCHEM, 47, 2012
|
|
3NXY
 
 | | Preferential Selection of Isomer Binding from Chiral Mixtures: Alernate Binding Modes Observed fro the E- and Z-isomers of a Series of 5-Substituted 2,4-Diaminofuro[2,3-d]pyrimidines as Ternary Complexes with NADPH and Human Dihydrofolate Reductase | | Descriptor: | 5-[(1E,3R)-2-(2-methoxyphenyl)-3-methylpent-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V. | | Deposit date: | 2010-07-14 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Preferential selection of isomer binding from chiral mixtures: alternate binding modes observed for the E and Z isomers of a series of 5-substituted 2,4-diaminofuro[2,3-d]pyrimidines as ternary complexes with NADPH and human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2BB7
 
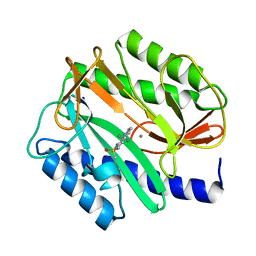 | |
1F9E
 
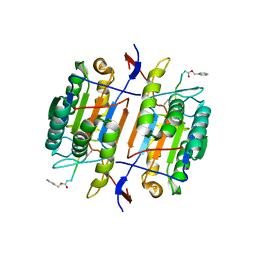 | | CASPASE-8 SPECIFICITY PROBED AT SUBSITE S4: CRYSTAL STRUCTURE OF THE CASPASE-8-Z-DEVD-CHO | | Descriptor: | (PHQ)DEVD, Caspase-8 subunit p10, Caspase-8 subunit p18 | | Authors: | Blanchard, H, Donepudi, M, Tschopp, M, Kodandapani, L, Wu, J.C, Grutter, M.G. | | Deposit date: | 2000-07-10 | | Release date: | 2001-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Caspase-8 specificity probed at subsite S(4): crystal structure of the caspase-8-Z-DEVD-cho complex.
J.Mol.Biol., 302, 2000
|
|
4K7M
 
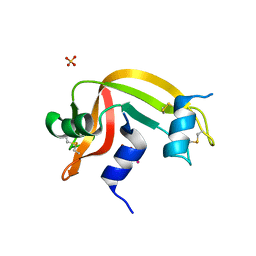 | | Crystal structure of RNase S variant (K7C/Q11C) with bound mercury ions | | Descriptor: | MERCURY (II) ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Genz, M, Straeter, N. | | Deposit date: | 2013-04-17 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Apo- and Metalated Thiolate containing RNase S as Structural Basis for the Design of Artificial Metalloenzymes by Peptide- Protein Complementation
Z.Anorg.Allg.Chem., 639, 2013
|
|
3OOE
 
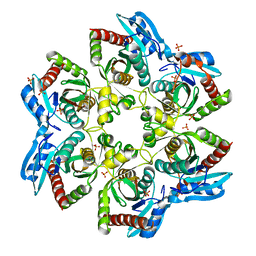 | | Crystal structure of E. Coli purine nucleoside phosphorylase with PO4 | | Descriptor: | PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | Authors: | Mikleusevic, G, Stefanic, Z, Narzyk, M, Wielgus-Kutrowska, B, Bzowska, A, Luic, M. | | Deposit date: | 2010-08-31 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Validation of the catalytic mechanism of Escherichia coli purine nucleoside phosphorylase by structural and kinetic studies.
Biochimie, 93, 2011
|
|
2GUF
 
 | | In meso crystal structure of the cobalamin transporter, BtuB | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FORMIC ACID, Vitamin B12 transporter btuB, ... | | Authors: | Caffrey, M, Cherezov, V, Yamashita, E, Cramer, W.A. | | Deposit date: | 2006-04-29 | | Release date: | 2006-12-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In Meso Structure of the Cobalamin Transporter, BtuB, at 1.95 A Resolution.
J.Mol.Biol., 364, 2006
|
|
2H8I
 
 | |
3OPV
 
 | | Crystal structure of E. Coli purine nucleoside phosphorylase Arg24Ala mutant | | Descriptor: | PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | Authors: | Mikleusevic, G, Stefanic, Z, Narzyk, M, Wielgus-Kutrowska, B, Bzowska, A, Luic, M. | | Deposit date: | 2010-09-02 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Validation of the catalytic mechanism of Escherichia coli purine nucleoside phosphorylase by structural and kinetic studies.
Biochimie, 93, 2011
|
|
2AMD
 
 | | Crystal Structure Of SARS_CoV Mpro in Complex with an Inhibitor N9 | | Descriptor: | 3C-like proteinase, N-(3-FUROYL)-D-VALYL-L-VALYL-N~1~-((1R,2Z)-4-ETHOXY-4-OXO-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-D-LEUCINAMIDE | | Authors: | Yang, H, Xue, X, Yang, K, Zhao, Q, Bartlam, M, Rao, Z. | | Deposit date: | 2005-08-09 | | Release date: | 2005-09-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases.
Plos Biol., 3, 2005
|
|
