7LZD
 
 | | Crystal Structure of SETD2 bound to Compound 35 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase SETD2, ... | | Authors: | Farrow, N.A, Boriack-Sjodin, P. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a First-in-Class Inhibitor of the Histone Methyltransferase SETD2 Suitable for Preclinical Studies.
Acs Med.Chem.Lett., 12, 2021
|
|
7LZF
 
 | | Crystal Structure of SETD2 bound to Compound 57 | | Descriptor: | 1,2-ETHANEDIOL, 4-fluoro-N-[(1R,3S)-3-{(3S)-3-[(methanesulfonyl)(methyl)amino]pyrrolidin-1-yl}cyclohexyl]-7-methyl-1H-indole-2-carboxamide, Histone-lysine N-methyltransferase SETD2, ... | | Authors: | Farrow, N.A, Boriack-Sjodin, P. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery of a First-in-Class Inhibitor of the Histone Methyltransferase SETD2 Suitable for Preclinical Studies.
Acs Med.Chem.Lett., 12, 2021
|
|
6SMS
 
 | | Vegetative Insecticidal Protein 1 (Vip1Ac1) from Bacillus thuringiensis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SULFATE ION, ... | | Authors: | Rizkallah, P.J, Al-Maslookhi, H.S, Berry, C, Jones, D. | | Deposit date: | 2019-08-22 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bacillus thuringiensis insecticidal protein 1Ac, VIP1Ac
Not Published
|
|
5FYO
 
 | | Calcium-dependent phosphoinositol-specific phospholipase C from a Gram-negative bacterium, Pseudomonas sp, apo form, crystal form 1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PHOSPHOINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Norgaard, A, Segura, D.R, Blicher, T.H, Wilson, K.S. | | Deposit date: | 2016-03-08 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of a calcium-dependent phosphoinositide-specific phospholipase C from Pseudomonas sp. 62186, the first from a Gram-negative bacterium.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5FUL
 
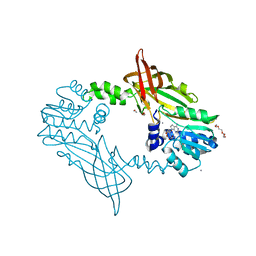 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with SAH | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-01-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
FEBS J., 284, 2017
|
|
5LRF
 
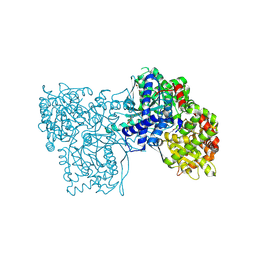 | | Crystal structure of Glycogen Phosphorylase b in complex with KS389 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(3-naphthalen-2-yl-1~{H}-1,2,4-triazol-5-yl)oxane-3,4,5-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kantsadi, A.L, Leonidas, D.D. | | Deposit date: | 2016-08-18 | | Release date: | 2017-06-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | van der Waals interactions govern C-beta-d-glucopyranosyl triazoles' nM inhibitory potency in human liver glycogen phosphorylase.
J. Struct. Biol., 199, 2017
|
|
7M1V
 
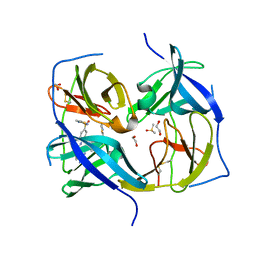 | | Structure of Zika virus NS2b-NS3 protease mutant binding the compound NSC86314 in the super-open conformation | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-{2,4-diamino-5-[2-(4-{[(2E)-1,3-thiazolidin-2-ylidene]sulfamoyl}phenyl)hydrazinyl]phenyl}hydrazinyl)-N-[(2S)-1,3-thiazolidin-2-yl]benzene-1-sulfonamide, CHLORIDE ION, ... | | Authors: | Aleshin, A.E, Shiryaev, S.A, Liddington, R.C. | | Deposit date: | 2021-03-15 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | To be provided
To Be Published
|
|
5LSA
 
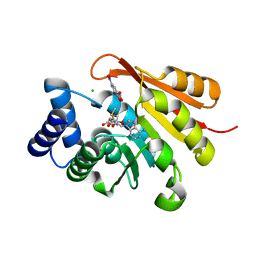 | |
7LNI
 
 | | SeMet CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Clostridioides difficile specific DNA adenine methyltransferase CamA squeezes and flips adenine out of DNA helix.
Nat Commun, 12, 2021
|
|
7LT5
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Cofactor SAH | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-02-18 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Clostridioides difficile specific DNA adenine methyltransferase CamA squeezes and flips adenine out of DNA helix.
Nat Commun, 12, 2021
|
|
5M2V
 
 | | Structure of GluK1 ligand-binding domain (S1S2) in complex with (2S,4R)-4-(2-carboxyphenoxy)pyrrolidine-2-carboxylic acid at 3.18 A resolution | | Descriptor: | (2~{S},4~{R})-4-(2-carboxyphenoxy)pyrrolidine-2-carboxylic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Frydenvang, K, Kastrup, J.S, Kristensen, C.M. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Design and Synthesis of a Series of l-trans-4-Substituted Prolines as Selective Antagonists for the Ionotropic Glutamate Receptors Including Functional and X-ray Crystallographic Studies of New Subtype Selective Kainic Acid Receptor Subtype 1 (GluK1) Antagonist (2S,4R)-4-(2-Carboxyphenoxy)pyrrolidine-2-carboxylic Acid.
J. Med. Chem., 60, 2017
|
|
4UZ9
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM - CRYSTAL FORM VII - SOS COMPLEX - 2.2A | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2014-09-04 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Notum Deacylates Wnt Proteins to Suppress Signalling Activity.
Nature, 519, 2015
|
|
4V0C
 
 | |
4V0H
 
 | | Human metallo beta lactamase domain containing protein 1 (hMBLAC1) | | Descriptor: | FE (III) ION, GLYCEROL, METALLO-BETA-LACTAMASE DOMAIN-CONTAINING PROTEIN 1 1 | | Authors: | Pettinati, I, McDonough, M.A, Brem, J, Schofield, C.J. | | Deposit date: | 2014-09-16 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Biosynthesis of histone messenger RNA employs a specific 3' end endonuclease.
Elife, 7, 2018
|
|
4V0Z
 
 | |
1DL2
 
 | | CRYSTAL STRUCTURE OF CLASS I ALPHA-1,2-MANNOSIDASE FROM SACCHAROMYCES CEREVISIAE AT 1.54 ANGSTROM RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CLASS I ALPHA-1,2-MANNOSIDASE, ... | | Authors: | Vallee, F, Lipari, F, Yip, P, Herscovics, A, Howell, P.L. | | Deposit date: | 1999-12-08 | | Release date: | 2000-02-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of a class I alpha1,2-mannosidase involved in N-glycan processing and endoplasmic reticulum quality control.
EMBO J., 19, 2000
|
|
7LPE
 
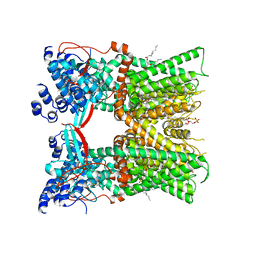 | | Cryo-EM structure of full-length TRPV1 with capsaicin at 48 degrees Celsius, in an open state, class 1 | | Descriptor: | (6E)-N-(4-hydroxy-3-methoxybenzyl)-8-methylnon-6-enamide, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Kwon, D.H, Zhang, F, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Heat-dependent opening of TRPV1 in the presence of capsaicin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LVS
 
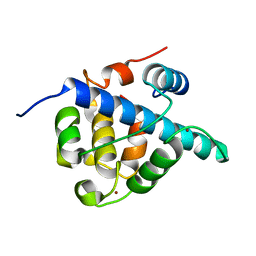 | | The CBP TAZ1 Domain in Complex with a CITED2-HIF-1-Alpha Fusion Peptide | | Descriptor: | Cbp/p300-interacting transactivator 2,Hypoxia-inducible factor 1-alpha, Histone lysine acetyltransferase CREBBP, ZINC ION | | Authors: | Appling, F.D, Berlow, R.B, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2021-02-26 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The molecular basis of allostery in a facilitated dissociation process.
Structure, 29, 2021
|
|
7LP2
 
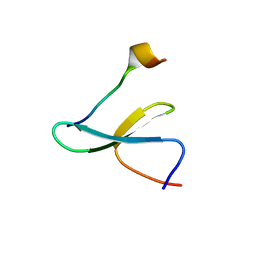 | | Structure of Nedd4L WW3 domain | | Descriptor: | Angiomotin, E3 ubiquitin-protein ligase, GLYCEROL, ... | | Authors: | Alian, A, Alam, S.L, Thompson, T, Rheinemann, L, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
4UWT
 
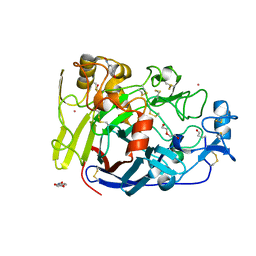 | | Hypocrea jecorina Cel7A E212Q mutant in complex with p-nitrophenyl cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE CEL7A, COBALT (II) ION, ... | | Authors: | Nutt, A, Momeni, M.H, Johansson, G, Stahlberg, J. | | Deposit date: | 2014-08-14 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Enzyme kinetics by GH7 cellobiohydrolases on chromogenic substrates is dictated by non-productive binding: insights from crystal structures and MD simulation.
Febs J., 2022
|
|
1DBO
 
 | | CRYSTAL STRUCTURE OF CHONDROITINASE B | | Descriptor: | 4-deoxy-alpha-D-glucopyranose-(1-3)-[beta-D-glucopyranose-(1-4)]2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, 4-deoxy-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, CHONDROITINASE B | | Authors: | Huang, W, Matte, A, Li, Y, Kim, Y.S, Linhardt, R.J, Su, H, Cygler, M. | | Deposit date: | 1999-11-03 | | Release date: | 2000-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 A resolution.
J.Mol.Biol., 294, 1999
|
|
7LMI
 
 | | SARS-CoV-1 3CLPro in complex with N-(4-(1H-pyrazol-4-yl)phenyl)-2-(1H-benzo[d][1,2,3]triazol-1-yl)-N-(thiophen-3-ylmethyl)acetamide | | Descriptor: | 2-(benzotriazol-1-yl)-~{N}-[4-(1~{H}-pyrazol-4-yl)phenyl]-~{N}-(thiophen-3-ylmethyl)ethanamide, 3C-like proteinase, GLYCEROL | | Authors: | Arya, T, Goins, C.M, Macdonald, J.D, Stauffer, S.R. | | Deposit date: | 2021-02-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Structure-Based Optimization of ML300-Derived, Noncovalent Inhibitors Targeting the Severe Acute Respiratory Syndrome Coronavirus 3CL Protease (SARS-CoV-2 3CL pro ).
J.Med.Chem., 65, 2022
|
|
7LHV
 
 | | Structure of Arabidopsis thaliana sulfate transporter AtSULTR4;1 | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, SULFATE ION, ... | | Authors: | Wang, L, Chen, K, Zhou, M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-08-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structure and function of an Arabidopsis thaliana sulfate transporter.
Nat Commun, 12, 2021
|
|
1DBG
 
 | | CRYSTAL STRUCTURE OF CHONDROITINASE B | | Descriptor: | 4-deoxy-alpha-D-glucopyranose-(1-3)-[beta-D-glucopyranose-(1-4)]2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, CHONDROITINASE B | | Authors: | Huang, W, Matte, A, Li, Y, Kim, Y.S, Linhardt, R.J, Su, H, Cygler, M. | | Deposit date: | 1999-11-02 | | Release date: | 2000-01-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 A resolution.
J.Mol.Biol., 294, 1999
|
|
5M9M
 
 | | Human angiogenin PD variant Q77P | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Angiogenin, ... | | Authors: | Bradshaw, W.J, Rehman, S, Pham, T.T.K, Thiyagarajan, N, Lee, R.L, Subramanian, V, Acharya, K.R. | | Deposit date: | 2016-11-01 | | Release date: | 2017-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into human angiogenin variants implicated in Parkinson's disease and Amyotrophic Lateral Sclerosis.
Sci Rep, 7, 2017
|
|
