1N8O
 
 | |
2ESU
 
 | | Crystal structure of Asn to Gln mutant of Winged Bean Chymotrypsin Inhibitor protein | | Descriptor: | Chymotrypsin inhibitor 3, NICKEL (II) ION, SULFATE ION | | Authors: | Dattagupta, J.K, Sen, U, Dasgupta, J, Khamrui, S. | | Deposit date: | 2005-10-27 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Spacer Asn Determines the Fate of Kunitz (STI) Inhibitors, as Revealed by Structural and Biochemical Studies on WCI Mutants.
Biochemistry, 45, 2006
|
|
3T62
 
 | | Crystal structure of recombinant Kunitz Type serine protease Inhibitor-1 from the Caribbean Sea anemone Stichodactyla helianthus in complex with bovine chymotrypsin | | Descriptor: | Chymotrypsinogen A, Kunitz-type proteinase inhibitor SHPI-1, SULFATE ION | | Authors: | Garcia-Fernandez, R, Dominguez, R, Oberthuer, D, Pons, T, Gonzalez-Gonzalez, Y, Chavez, M.A, Betzel, C, Redecke, L. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into chymotrypsin inhibition by the Kunitz-type inhibitor-1 from the marine invertebrate Stichodactyla helianthus
To be Published
|
|
1WKD
 
 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-08-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase reveal aspartate 102 as the active site nucleophile.
Biochemistry, 35, 1996
|
|
1WKE
 
 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-08-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase reveal aspartate 102 as the active site nucleophile.
Biochemistry, 35, 1996
|
|
1WKF
 
 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-08-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase reveal aspartate 102 as the active site nucleophile.
Biochemistry, 35, 1996
|
|
4DG1
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with polymorphism mutation K172A and K173A | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Tu, X, Kirby, K.A, Marchand, B, Sarafianos, S.G. | | Deposit date: | 2012-01-24 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | HIV-1 Reverse Transcriptase (RT) Polymorphism 172K Suppresses the Effect of Clinically Relevant Drug Resistance Mutations to Both Nucleoside and Non-nucleoside RT Inhibitors.
J.Biol.Chem., 287, 2012
|
|
9AVN
 
 | | Crystal Structure of CARD9 coiled-coil K156-K214 bound to Compound 1 | | Descriptor: | (2P)-2-(1-benzyl-3-phenyl-1H-pyrazol-4-yl)-5-chloro-1-[(1R)-1-cyclohexyl-2-(methylamino)-2-oxoethyl]-1H-1,3-benzimidazole-6-carboxylic acid, Caspase recruitment domain-containing protein 9 | | Authors: | Raymond, D.D, Lemke, C.T. | | Deposit date: | 2024-03-04 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.726 Å) | | Cite: | Human genetics guides the discovery of novel CARD9 inhibitors with anti-inflammatory activity in vitro as well as in vivo
To be published
|
|
9AVM
 
 | | Crystal Structure of CARD9 coiled-coil K156-K214 | | Descriptor: | Caspase recruitment domain-containing protein 9 | | Authors: | Raymond, D.D, Lemke, C.T. | | Deposit date: | 2024-03-04 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Human genetics guides the discovery of novel CARD9 inhibitors with anti-inflammatory activity in vitro as well as in vivo
To be published
|
|
5XSF
 
 | |
4V9E
 
 | |
4KKM
 
 | | Crystal structure of a FPP/GFPP synthase (Target EFI-501952) from Zymomonas mobilis, apo structure | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-05-06 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a FPP/GFPP synthase (Target EFI-501952) from Zymomonas mobilis, apo structure
To be Published
|
|
5IBU
 
 | | 6652 Fab (unbound) | | Descriptor: | 6652 Heavy Chain, 6652 Light Chain | | Authors: | Raymond, D.D, Harrison, S.C. | | Deposit date: | 2016-02-22 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Influenza immunization elicits antibodies specific for an egg-adapted vaccine strain.
Nat. Med., 22, 2016
|
|
5IBT
 
 | | UCA Fab (unbound) from 6515 Lineage | | Descriptor: | GLYCEROL, UCA Heavy Chain, UCA Light Chain | | Authors: | Raymond, D.D, Harrison, S.C. | | Deposit date: | 2016-02-22 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Influenza immunization elicits antibodies specific for an egg-adapted vaccine strain.
Nat. Med., 22, 2016
|
|
5IBL
 
 | |
4BK9
 
 | |
3SM0
 
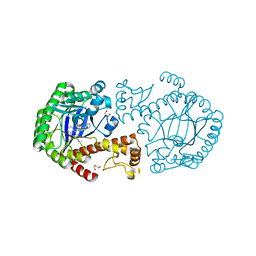 | | tRNA-Guanine Transglycosylase in complex with lin-Benzohypoxanthine Inhibitor | | Descriptor: | 4-{2-[(cyclopentylmethyl)amino]ethyl}-2-(methylamino)-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Klebe, G, Immekus, F, Heine, A. | | Deposit date: | 2011-06-27 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | From lin-Benzoguanines to lin-Benzohypoxanthines as Ligands for Zymomonas mobilis tRNA-Guanine Transglycosylase: Replacement of Protein-Ligand Hydrogen Bonding by Importing Water Clusters.
Chemistry, 18, 2012
|
|
3UPS
 
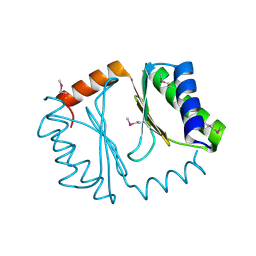 | |
3RR4
 
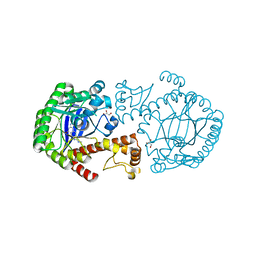 | | tRNA-Guanine Transglycosylase in complex with N-Methyl-lin-Benzoguanine Inhibitor | | Descriptor: | 2,6-bis(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Klebe, G, Immekus, F, Heine, A. | | Deposit date: | 2011-04-29 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | From lin-Benzoguanines to lin-Benzohypoxanthines as Ligands for Zymomonas mobilis tRNA-Guanine Transglycosylase: Replacement of Protein-Ligand Hydrogen Bonding by Importing Water Clusters.
Chemistry, 18, 2012
|
|
1YXA
 
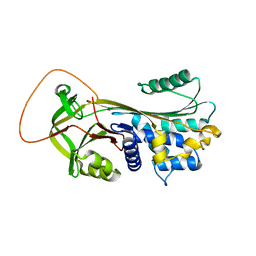 | | Serpina3n, a murine orthologue of human antichymotrypsin | | Descriptor: | serine (or cysteine) proteinase inhibitor, clade A, member 3N | | Authors: | Horvath, A.J, Irving, J.A, Law, R.H, Rossjohn, J, Bottomley, S.P, Quinsey, N.S, Pike, R.N, Coughlin, P.B, Whisstock, J.C. | | Deposit date: | 2005-02-20 | | Release date: | 2005-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The murine orthologue of human antichymotrypsin: a structural paradigm for clade A3 serpins.
J.Biol.Chem., 280, 2005
|
|
2YPK
 
 | | Structural features underlying T-cell receptor sensitivity to concealed MHC class I micropolymorphisms | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, B-57 ALPHA CHAIN, ... | | Authors: | Stewart-Jones, G.B, Simpson, P, Van Der Merwe, P.A, Easterbrook, P, Mcmichael, A.J, Rowland-Jones, S.L, Jones, E.Y, Gillespie, G.M. | | Deposit date: | 2012-10-30 | | Release date: | 2012-11-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Features Underlying T-Cell Receptor Sensitivity to Concealed Mhc Class I Micropolymorphisms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TLL
 
 | | tRNA-Guanine Transglycosylase in complex with N-Ethyl-lin-benzoguanine Inhibitor | | Descriptor: | 6-(ethylamino)-2-(methylamino)-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Klebe, G, Immekus, F, Heine, A. | | Deposit date: | 2011-08-30 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | From lin-Benzoguanines to lin-Benzohypoxanthines as Ligands for Zymomonas mobilis tRNA-Guanine Transglycosylase: Replacement of Protein-Ligand Hydrogen Bonding by Importing Water Clusters.
Chemistry, 18, 2012
|
|
5W6G
 
 | | Human antibody 6649 in complex with influenza hemagglutinin H1 Solomon Islands | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6649 antibody heavy chain, ... | | Authors: | Raymond, D.D, Harrison, S.C. | | Deposit date: | 2017-06-16 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5W6C
 
 | | UCA Fab (unbound) from 6649 Lineage | | Descriptor: | UCA Fab heavy chain, UCA Fab light chain | | Authors: | Raymond, D.D, Harrison, S.C. | | Deposit date: | 2017-06-16 | | Release date: | 2017-12-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.634 Å) | | Cite: | Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4MYE
 
 | | Cymosema roseum seed lectin structure complexed with X-man | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, Cymbosema roseum mannose-specific lectin, ... | | Authors: | Barroso-Neto, I.L, Rocha, B.A.M, Teixeira, C.S, Santiago, M.Q, Souza, L.A.G, Pires, A.F, Assreuy, A.M.S, Delatorre, P, Cavada, B.S. | | Deposit date: | 2013-09-27 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cymosema roseum seed lectin structure complexed with X-man
To be Published
|
|
