8OJO
 
 | | Galectin-3 in complex with 2,6-Anhydro-5-S-(beta-D-galactopyranosyl)-5-thio-D-altritol | | Descriptor: | 1-deoxy-alpha-D-mannopyranose, 1-thio-beta-D-galactopyranose, Galectin-3, ... | | Authors: | Tsagkarakou, A.S, Leonidas, D.D. | | Deposit date: | 2023-03-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Strong Binding of C -Glycosylic1,2-Thiodisaccharides to Galectin-3─Enthalpy-Driven Affinity Enhancement by Water-Mediated Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
7Q22
 
 | | cryo iDPC-STEM structure recorded with CSA 2.0 | | Descriptor: | Capsid protein, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Sachse, C, Leidl, M.L. | | Deposit date: | 2021-10-22 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Single-particle cryo-EM structures from iDPC-STEM at near-atomic resolution.
Nat.Methods, 19, 2022
|
|
7Q2Q
 
 | | cryo iDPC-STEM structure recorded with CSA 3.5 | | Descriptor: | Capsid protein, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Sachse, C, Leidl, M.L. | | Deposit date: | 2021-10-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Single-particle cryo-EM structures from iDPC-STEM at near-atomic resolution.
Nat.Methods, 19, 2022
|
|
7Q2S
 
 | | cryo iDPC-STEM structure recorded with CSA 4.5 | | Descriptor: | Capsid protein, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Sachse, C, Leidl, M.L. | | Deposit date: | 2021-10-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Single-particle cryo-EM structures from iDPC-STEM at near-atomic resolution.
Nat.Methods, 19, 2022
|
|
8EK4
 
 | |
7Q23
 
 | | cryo iDPC-STEM structure recorded with CSA 3.0 | | Descriptor: | Capsid protein, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Sachse, C, Leidl, M.L. | | Deposit date: | 2021-10-22 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Single-particle cryo-EM structures from iDPC-STEM at near-atomic resolution.
Nat.Methods, 19, 2022
|
|
7Q2R
 
 | | cryo iDPC-STEM structure recorded with CSA 4.0 | | Descriptor: | Capsid protein, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Sachse, C, Leidl, M.L. | | Deposit date: | 2021-10-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Single-particle cryo-EM structures from iDPC-STEM at near-atomic resolution.
Nat.Methods, 19, 2022
|
|
7BML
 
 | |
7BR5
 
 | | Lysozyme-sugar complex in H2O | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Tanaka, I, Chatake, T. | | Deposit date: | 2020-03-26 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Recent structural insights into the mechanism of lysozyme hydrolysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7C03
 
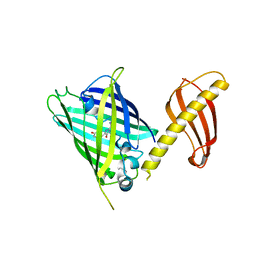 | | Crystal structure of POLArISact(T57S), genetically encoded probe for fluorescent polarization | | Descriptor: | POLArISact(T57S) | | Authors: | Tomabechi, Y, Sakai, N, Shirouzu, M. | | Deposit date: | 2020-04-30 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | POLArIS, a versatile probe for molecular orientation, revealed actin filaments associated with microtubule asters in early embryos.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7PO7
 
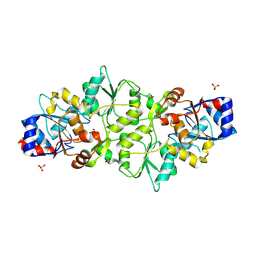 | | Phosphoglycolate phosphatase from Mus musculus | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Schloetzer, J, Schindelin, H, Fratz, S. | | Deposit date: | 2021-09-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Glycolytic flux control by drugging phosphoglycolate phosphatase.
Nat Commun, 13, 2022
|
|
7POE
 
 | | Phosphoglycolate Phosphatase with Inhibitor CP1 | | Descriptor: | 2-[[4-[4-[(2-carboxyphenyl)carbamoyl]phenoxy]phenyl]carbonylamino]benzoic acid, GLYCEROL, Glycerol-3-phosphate phosphatase, ... | | Authors: | Schloetzer, J, Fratz, S, Schindelin, H. | | Deposit date: | 2021-09-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Glycolytic flux control by drugging phosphoglycolate phosphatase.
Nat Commun, 13, 2022
|
|
8EWV
 
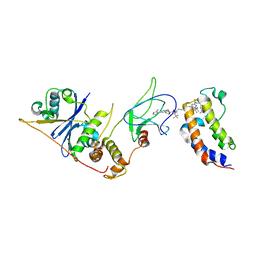 | | DNA-encoded library (DEL)-enabled discovery of proximity inducing small molecules | | Descriptor: | Bromodomain-containing protein 4, Elongin-B, Elongin-C, ... | | Authors: | Schreiber, S.L, Shu, W, Ma, X, Michaud, G, Bonazzi, S, Berst, F. | | Deposit date: | 2022-10-24 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | DNA-encoded library-enabled discovery of proximity-inducing small molecules.
Nat.Chem.Biol., 20, 2024
|
|
8ETQ
 
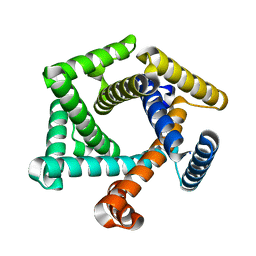 | |
8PPN
 
 | |
1KB5
 
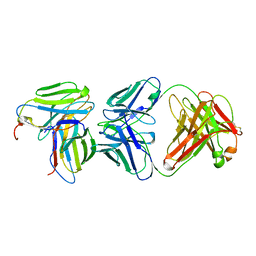 | | MURINE T-CELL RECEPTOR VARIABLE DOMAIN/FAB COMPLEX | | Descriptor: | ANTIBODY DESIRE-1, KB5-C20 T-CELL ANTIGEN RECEPTOR | | Authors: | Housset, D, Mazza, G, Gregoire, C, Piras, C, Malissen, B, Fontecilla-Camps, J.C. | | Deposit date: | 1997-04-06 | | Release date: | 1998-04-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The three-dimensional structure of a T-cell antigen receptor V alpha V beta heterodimer reveals a novel arrangement of the V beta domain.
EMBO J., 16, 1997
|
|
7Q5I
 
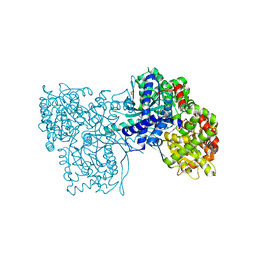 | | A glucose-based molecular rotor probes the catalytic site of glycogen phosphorylase. | | Descriptor: | 2-cyano-3-[4-(dimethylamino)phenyl]-~{N}-[(2~{R},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]propanamide, BETA-MERCAPTOETHANOL, CARBONATE ION, ... | | Authors: | Neofytos, D.D, Chrysina, E.D. | | Deposit date: | 2021-11-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A glucose-based molecular rotor inhibitor of glycogen phosphorylase as a probe of cellular enzymatic function.
Org.Biomol.Chem., 20, 2022
|
|
8FTG
 
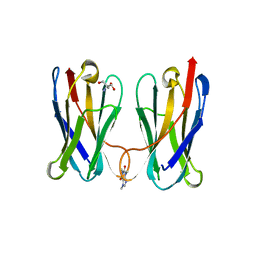 | | Biophysical and Structural Characterization of an Anti-Caffeine VHH Antibody | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Anti-Caffeine VHH Antibody, CAFFEINE, ... | | Authors: | Horn, J.R, Smith, C.A, Sonneson, G.J, Walter, R. | | Deposit date: | 2023-01-12 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Molecular recognition requires dimerization of a VHH antibody.
Mabs, 15, 2023
|
|
7CGC
 
 | |
7CGD
 
 | | Silver-bound E.coli malate dehydrogenase | | Descriptor: | Malate dehydrogenase, SILVER ION | | Authors: | Wang, H, Wang, M, Sun, H. | | Deposit date: | 2020-07-01 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Atomic differentiation of silver binding preference in protein targets: Escherichia coli malate dehydrogenase as a paradigm.
Chem Sci, 11, 2020
|
|
7CGV
 
 | | Full consensus L-threonine 3-dehydrogenase, FcTDH-IIYM (NAD+ bound form) | | Descriptor: | Artificial L-threonine 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Motoyama, T, Hiramatsu, N, Asano, Y, Nakano, S, Ito, S. | | Deposit date: | 2020-07-02 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Protein Sequence Selection Method That Enables Full Consensus Design of Artificial l-Threonine 3-Dehydrogenases with Unique Enzymatic Properties.
Biochemistry, 59, 2020
|
|
7CND
 
 | | NCI-1 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, CRM1 isoform 1, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-07-31 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of the First Noncovalent Small-Molecule Inhibitor of CRM1.
J.Med.Chem., 64, 2021
|
|
8GVM
 
 | | The structure of azide-bound cytochrome C oxidase determined using the crystals exposed to 20 mm azide solution for 4 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimada, A. | | Deposit date: | 2022-09-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity
J. Biol. Chem., 293, 2018
|
|
8GFT
 
 | | Hsp90 provides platform for CRaf dephosphorylation by PP5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, ... | | Authors: | Jaime-Garza, M, Nowotny, C.A, Coutandin, D, Wang, F, Tabios, M, Agard, D.A. | | Deposit date: | 2023-03-08 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Hsp90 provides a platform for kinase dephosphorylation by PP5.
Nat Commun, 14, 2023
|
|
8QFW
 
 | | Murine pyridoxal phosphatase in complex with 7,8-dihydroxyflavone | | Descriptor: | 7,8-bis(oxidanyl)-2-phenyl-chromen-4-one, CITRIC ACID, Chronophin, ... | | Authors: | Schindelin, H, Gohla, A. | | Deposit date: | 2023-09-05 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 7,8-Dihydroxyflavone is a direct inhibitor of human and murine pyridoxal phosphatase.
Elife, 13, 2024
|
|
