3CDJ
 
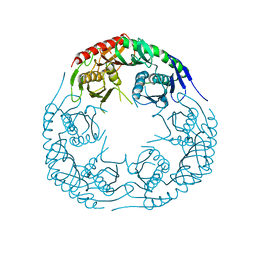 | | Crystal structure of the E. coli KH/S1 domain truncated PNPase | | Descriptor: | Polynucleotide phosphorylase | | Authors: | Shi, Z, Yang, W.Z, Lin-Chao, S, Chak, K.F, Yuan, H.S. | | Deposit date: | 2008-02-27 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive RNA degradation.
Rna, 14, 2008
|
|
3CDI
 
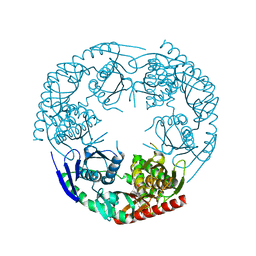 | | Crystal structure of E. coli PNPase | | Descriptor: | Polynucleotide phosphorylase | | Authors: | Shi, Z, Yang, W.Z, Lin-Chao, S, Chak, K.F, Yuan, H.S. | | Deposit date: | 2008-02-27 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive RNA degradation.
Rna, 14, 2008
|
|
3CFZ
 
 | |
2QU4
 
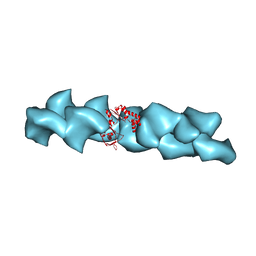 | | Model for Bacterial ParM Filament | | Descriptor: | Plasmid segregation protein parM | | Authors: | Orlova, A, Garner, E.C, Galkin, V.E, Heuser, J, Mullins, R.D, Egelman, E.H. | | Deposit date: | 2007-08-03 | | Release date: | 2007-09-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | The structure of bacterial ParM filaments.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2QOM
 
 | | The crystal structure of the E.coli EspP autotransporter Beta-domain. | | Descriptor: | Serine protease espP | | Authors: | Barnard, T.J, Dautin, N, Lukacik, P, Bernstein, H.D, Buchanan, S.K. | | Deposit date: | 2007-07-20 | | Release date: | 2007-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Autotransporter structure reveals intra-barrel cleavage followed by conformational changes.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2QRI
 
 | |
2QRT
 
 | |
5JM4
 
 | | Crystal structure of 14-3-3zeta in complex with a cyclic peptide involving an adamantyl and a dicarboxy side chain | | Descriptor: | 14-3-3 protein zeta/delta, BENZOIC ACID, GLN-GLY-MKD-ANG-ASP-MKD-LEU-ASP-LEU-ALA-CLU | | Authors: | Bier, D, Krueger, D, Glas, A, Wallraven, K, Ottmann, C, Hennig, S, Grossmann, T. | | Deposit date: | 2016-04-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure-Based Design of Non-natural Macrocyclic Peptides That Inhibit Protein-Protein Interactions.
J. Med. Chem., 60, 2017
|
|
3SQ6
 
 | | Crystal Structures of the Ligand Binding Domain of a Pentameric Alpha7 Nicotinic Receptor Chimera with its Agonist Epibatidine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EPIBATIDINE, ... | | Authors: | Li, S.-X, Huang, S, Bren, N, Noridomi, K, Dellisanti, C, Sine, S, Chen, L. | | Deposit date: | 2011-07-05 | | Release date: | 2011-09-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ligand-binding domain of an alpha 7-nicotinic receptor chimera and its complex with agonist.
Nat.Neurosci., 14, 2011
|
|
3T4N
 
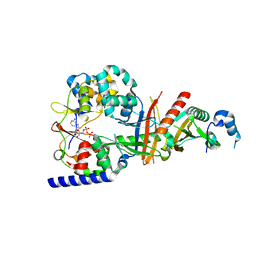 | | Structure of the regulatory fragment of Saccharomyces cerevisiae AMPK in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmdit, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-07-26 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
3SQ9
 
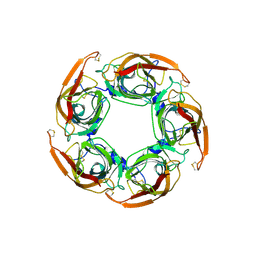 | | Crystal Structures of the Ligand Binding Domain of a Pentameric Alpha7 Nicotinic Receptor Chimera | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Neuronal acetylcholine receptor subunit alpha-7, ... | | Authors: | Li, S.-X, Huang, S, Bren, N, Noridomi, K, Dellisanti, C, Sine, S, Chen, L. | | Deposit date: | 2011-07-05 | | Release date: | 2011-09-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Ligand-binding domain of an alpha 7-nicotinic receptor chimera and its complex with agonist.
Nat.Neurosci., 14, 2011
|
|
2F31
 
 | |
2F3U
 
 | |
2F8T
 
 | |
2F9W
 
 | | Structure of the type III CoaA from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PANTOTHENOIC ACID, ... | | Authors: | Leonardi, R, Yun, M.K, Chohnan, S, White, S.W, Rock, C.O, Jackowski, S. | | Deposit date: | 2005-12-06 | | Release date: | 2006-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Prokaryotic Type II and Type III Pantothenate Kinases: The Same Monomer Fold Creates Dimers with Distinct Catalytic Properties.
Structure, 14, 2006
|
|
2F3S
 
 | |
2EYY
 
 | | CT10-Regulated Kinase isoform I | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2F1M
 
 | |
2F1K
 
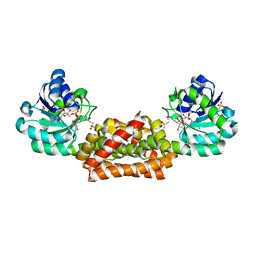 | | Crystal structure of Synechocystis arogenate dehydrogenase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, prephenate dehydrogenase | | Authors: | Legrand, P, Dumas, R, Seux, M, Rippert, P, Ravelli, R, Ferrer, J.-L, Matringe, M. | | Deposit date: | 2005-11-14 | | Release date: | 2006-05-09 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biochemical Characterization and Crystal Structure of Synechocystis Arogenate Dehydrogenase Provide Insights into Catalytic Reaction
Structure, 14, 2006
|
|
2F2U
 
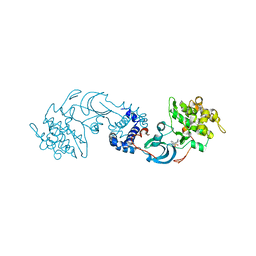 | | crystal structure of the Rho-kinase kinase domain | | Descriptor: | 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, Rho-associated protein kinase 2 | | Authors: | Yamaguchi, H, Hakoshima, T. | | Deposit date: | 2005-11-18 | | Release date: | 2006-04-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism for the regulation of rho-kinase by dimerization and its inhibition by fasudil
Structure, 14, 2006
|
|
2FMS
 
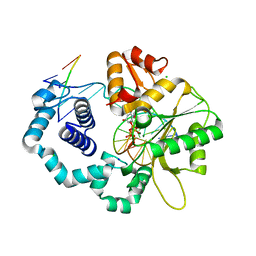 | | DNA Polymerase beta with a gapped DNA substrate and dUMPNPP with magnesium in the catalytic site | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Krahn, J.M, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Magnesium-induced assembly of a complete DNA polymerase catalytic complex.
Structure, 14, 2006
|
|
2FQL
 
 | |
2FR0
 
 | |
2FMQ
 
 | | Sodium in active site of DNA Polymerase Beta | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Krahn, J.M, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Magnesium-induced assembly of a complete DNA polymerase catalytic complex.
Structure, 14, 2006
|
|
2FYO
 
 | | Crystal structure of rat carnitine palmitoyltransferase 2 in space group P43212 | | Descriptor: | Carnitine O-palmitoyltransferase II, mitochondrial | | Authors: | Rufer, A.C, Thoma, R, Benz, J, Stihle, M, Gsell, B, De Roo, E, Banner, D.W, Mueller, F, Chomienne, O, Hennig, M. | | Deposit date: | 2006-02-08 | | Release date: | 2007-02-08 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of carnitine palmitoyltransferase 2 and implications for diabetes treatment
Structure, 14, 2006
|
|
