8IRA
 
 | | XFEL structure of cyanobacterial photosystem II following one flash (1F) with a 200-microsecond delay | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
5MBC
 
 | | Structure of a bacterial light-regulated adenylyl cylcase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
8C3J
 
 | | Stapled peptide SP2 in complex with humanised RadA mutant HumRadA22 | | Descriptor: | 2-[(4,6-diethyl-1,3,5-triazin-2-yl)-methyl-amino]ethanoic acid, Breast cancer type 2 susceptibility protein, DNA repair and recombination protein RadA | | Authors: | Pantelejevs, T, Hyvonen, M. | | Deposit date: | 2022-12-26 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | A recombinant approach for stapled peptide discovery yields inhibitors of the RAD51 recombinase.
Chem Sci, 14, 2023
|
|
5U90
 
 | | Crystal structure of Co-CAO1 in complex with resveratrol | | Descriptor: | COBALT (II) ION, Carotenoid oxygenase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Sui, X, Palczewski, k, Banerjee, S, Kiser, P.D. | | Deposit date: | 2016-12-15 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Spectroscopy of Alkene-Cleaving Dioxygenases Containing an Atypically Coordinated Non-Heme Iron Center.
Biochemistry, 56, 2017
|
|
5XVQ
 
 | | Crystal structure of monkey Nicotinamide N-methyltransferase (NNMT) bound with end product, 1-methyl Nicotinamide (MNA) | | Descriptor: | 3-carbamoyl-1-methylpyridin-1-ium, GLYCEROL, Nicotinamide N-methyltransferase (NNMT), ... | | Authors: | Birudukota, S, Swaminathan, S, Thakur, M.K, Parveen, R, Kandan, S, Kannt, A, Gosu, R. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structures of monkey and mouse nicotinamide N-methyltransferase (NNMT) bound with end product, 1-methyl nicotinamide
Biochem. Biophys. Res. Commun., 491, 2017
|
|
5U8W
 
 | | Dihydrolipoamide dehydrogenase (LpdG) from Pseudomonas aeruginosa bound to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, Dihydrolipoyl dehydrogenase, ... | | Authors: | Glasser, N.R, Wang, B.X, Hoy, J.A, Newman, D.K. | | Deposit date: | 2016-12-15 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Pyruvate and alpha-Ketoglutarate Dehydrogenase Complexes of Pseudomonas aeruginosa Catalyze Pyocyanin and Phenazine-1-carboxylic Acid Reduction via the Subunit Dihydrolipoamide Dehydrogenase.
J. Biol. Chem., 292, 2017
|
|
5XVV
 
 | | Crystal Structure of Forward Inhibited Aspergillus niger Glutamate Dehydrogenase With Both Apo- and Alpha Ketoglutarate Bound Subunits | | Descriptor: | 2-OXOGLUTARIC ACID, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Prakash, P, Punekar, N.S, Bhaumik, P. | | Deposit date: | 2017-06-28 | | Release date: | 2018-03-21 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the catalytic mechanism and alpha-ketoglutarate cooperativity of glutamate dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
5J7J
 
 | |
5XGX
 
 | | Crystal structure of colwellia psychrerythraea strain 34H isoaspartyl dipeptidase E80Q mutant complexed with beta-isoaspartyl lysine | | Descriptor: | D-ASPARTIC ACID, D-LYSINE, Isoaspartyl dipeptidase, ... | | Authors: | Lee, J.H, Lee, C.W, Park, S.H. | | Deposit date: | 2017-04-18 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure and functional characterization of an isoaspartyl dipeptidase (CpsIadA) from Colwellia psychrerythraea strain 34H.
PLoS ONE, 12, 2017
|
|
8HWO
 
 | |
3NHD
 
 | | GYVLGS segment 127-132 from human prion with V129 | | Descriptor: | ACETIC ACID, Major prion protein | | Authors: | Apostol, M.I, Eisenberg, D. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystallographic studies of prion protein (PrP) segments suggest how structural changes encoded by polymorphism at residue 129 modulate susceptibility to human prion disease.
J.Biol.Chem., 285, 2010
|
|
1IKF
 
 | | A CONFORMATION OF CYCLOSPORIN A IN AQUEOUS ENVIRONMENT REVEALED BY THE X-RAY STRUCTURE OF A CYCLOSPORIN-FAB COMPLEX | | Descriptor: | CYCLOSPORIN A, IGG1-KAPPA R45-45-11 FAB (HEAVY CHAIN), IGG1-KAPPA R45-45-11 FAB (LIGHT CHAIN) | | Authors: | Vix, O, Altschuh, D, Rees, B, Thierry, J.-C. | | Deposit date: | 1993-12-09 | | Release date: | 1995-03-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Conformation of Cyclosporin a in Aqueous Environment Revealed by the X-Ray Structure of a Cyclosporin-Fab Complex.
Science, 256, 1992
|
|
8I6Y
 
 | | Crystal structure of Arabidopsis thaliana LOX1 | | Descriptor: | FE (III) ION, Linoleate 9S-lipoxygenase 1 | | Authors: | Liu, X, Liu, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | UV-B light signal mediates stomatal closure by activating the 9-lipoxygenase pathway
To Be Published
|
|
5UBU
 
 | | 2.75 Angstrom Resolution Crystal Structure of Acetamidase from Yersinia enterocolitica. | | Descriptor: | Putative acetamidase/formamidase, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Flores, K, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom Resolution Crystal Structure of Acetamidase from Yersinia enterocolitica.
To Be Published
|
|
3SI5
 
 | | Kinetochore-BUBR1 kinase complex | | Descriptor: | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta, Protein CASC5 | | Authors: | Blundell, T.L, Chirgadze, D.Y, Bolanos-Garcia, V.M. | | Deposit date: | 2011-06-17 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a Blinkin-BUBR1 Complex Reveals an Interaction Crucial for Kinetochore-Mitotic Checkpoint Regulation via an Unanticipated Binding Site.
Structure, 19, 2011
|
|
5XKD
 
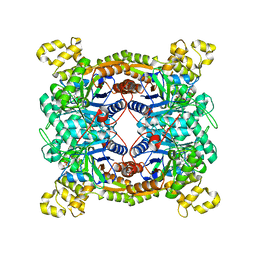 | | Crystal structure of dibenzothiophene sulfone monooxygenase BdsA in complex with FMN at 2.4 angstrom | | Descriptor: | Dibenzothiophene desulfurization enzyme A, FLAVIN MONONUCLEOTIDE | | Authors: | Gu, L, Su, T, Liu, S, Su, J. | | Deposit date: | 2017-05-07 | | Release date: | 2018-05-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural and Biochemical Characterization of BdsA fromBacillus subtilisWU-S2B, a Key Enzyme in the "4S" Desulfurization Pathway.
Front Microbiol, 9, 2018
|
|
5CMV
 
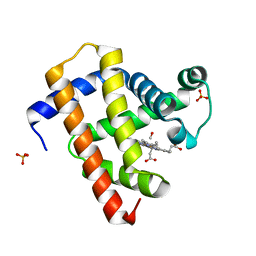 | | Ultrafast dynamics in myoglobin: dark-state, CO-ligated structure | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
1U0J
 
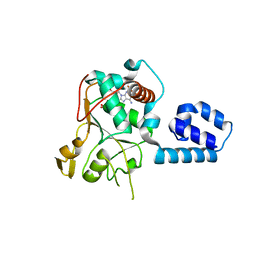 | | Crystal Structure of AAV2 Rep40-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication protein | | Authors: | James, J.A, Aggarwal, A.K, Linden, R.M, Escalante, C.R. | | Deposit date: | 2004-07-13 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of adeno-associated virus type 2 Rep40-ADP complex: Insight into
nucleotide recognition and catalysis by superfamily 3 helicases
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
5CND
 
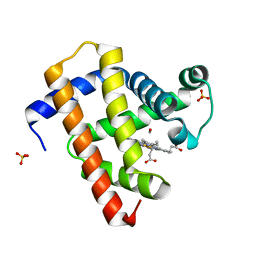 | | Ultrafast dynamics in myoglobin: 3 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
8IH7
 
 | | AmnG-AmnH complex | | Descriptor: | 4-hydroxy-2-oxovalerate aldolase, Acetaldehyde dehydrogenase, OXALATE ION, ... | | Authors: | Su, D, Shi, Q.L. | | Deposit date: | 2023-02-22 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | AmnG-AmnH complex
To Be Published
|
|
7YMI
 
 | | PSII-Pcb Dimer of Acaryochloris Marina | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Shen, L.L, Gao, Y.Z, Wang, W.D, Zhang, X, Shen, J.R, Wang, P.Y, Han, G.Y. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a large photosystem II supercomplex from Acaryochloris marina.
To Be Published
|
|
7YMM
 
 | | PSII-Pcb Tetramer of Acaryochloris Marina | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Shen, L.L, Gao, Y.Z, Wang, W.D, Zhang, X, Shen, J.R, Wang, P.Y, Han, G.Y. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a large photosystem II supercomplex from Acaryochloris marina.
To Be Published
|
|
8IL5
 
 | |
8CL6
 
 | | Lysozyme in matrix of hydroxyethylcellulose (HEC) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, LYSOZYME, ... | | Authors: | Bertrand, Q, Kepa, M.W, Weinert, T, Wranik, M, Standfuss, J. | | Deposit date: | 2023-02-16 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | A multi-reservoir extruder for time-resolved serial protein crystallography and compound screening at X-ray free-electron lasers.
Nat Commun, 14, 2023
|
|
5CN5
 
 | | Ultrafast dynamics in myoglobin: 0 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
