6AGS
 
 | |
1AGT
 
 | | SOLUTION STRUCTURE OF THE POTASSIUM CHANNEL INHIBITOR AGITOXIN 2: CALIPER FOR PROBING CHANNEL GEOMETRY | | Descriptor: | AGITOXIN 2 | | Authors: | Krezel, A.M, Kasibhatla, C, Hidalgo, P, Mackinnon, R, Wagner, G. | | Deposit date: | 1995-04-14 | | Release date: | 1995-07-10 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the potassium channel inhibitor agitoxin 2: caliper for probing channel geometry.
Protein Sci., 4, 1995
|
|
8BZD
 
 | | deAc-FC stabilizer of 14-3-3 and ERalpha | | Descriptor: | 14-3-3 protein sigma, De-acetylated Fusicoccin, ERalpha peptide, ... | | Authors: | Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-14 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Stabilization of the Estrogen receptor alpha - 14-3-3 interaction as a potential intervention strategy for endocrine resistance in breast cancer
To Be Published
|
|
1U0J
 
 | | Crystal Structure of AAV2 Rep40-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication protein | | Authors: | James, J.A, Aggarwal, A.K, Linden, R.M, Escalante, C.R. | | Deposit date: | 2004-07-13 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of adeno-associated virus type 2 Rep40-ADP complex: Insight into
nucleotide recognition and catalysis by superfamily 3 helicases
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8BZT
 
 | | FC-NAg stabilizer of 14-3-3 and ERalpha | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, MAGNESIUM ION, ... | | Authors: | Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-15 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stabilization of the Estrogen receptor alpha - 14-3-3 interaction as a potential intervention strategy for endocrine resistance in breast cancer
To Be Published
|
|
8BZH
 
 | | FC-NAc stabilizer of 14-3-3 and ERalpha | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, MAGNESIUM ION, ... | | Authors: | Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-14 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Stabilization of the Estrogen receptor alpha - 14-3-3 interaction as a potential intervention strategy for endocrine resistance in breast cancer
To Be Published
|
|
6AJG
 
 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with SQ109 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, DODECYL-BETA-D-MALTOSIDE, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
1AKB
 
 | |
3W1H
 
 | |
6AHL
 
 | | Crystal Structure of HEWL in complex with Cinnamaldehyde (in the aroma form) after 5 hours under fibrillation conditions | | Descriptor: | 4-CARBOXYCINNAMIC ACID, ACETATE ION, CHLORIDE ION, ... | | Authors: | Seraj, Z, Seyedarabi, A. | | Deposit date: | 2018-08-19 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cinnamaldehyde and Phenyl Ethyl Alcohol promote the entrapment of intermediate species of HEWL, as revealed by structural, kinetics and thermal stability studies.
Sci Rep, 9, 2019
|
|
1IY8
 
 | | Crystal Structure of Levodione Reductase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, LEVODIONE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sogabe, S, Fukami, T, Shiratori, Y, Yoshizumi, A, Wada, M. | | Deposit date: | 2002-07-25 | | Release date: | 2003-05-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure and Stereospecificity of Levodione Reductase from Corynebacterium aquaticum M-13
J.BIOL.CHEM., 278, 2003
|
|
8BZG
 
 | | FC-31 stabilizer of 14-3-3 and ERalpha | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, Fusicoccin 31, ... | | Authors: | Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-14 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Stabilization of the Estrogen receptor alpha - 14-3-3 interaction as a potential intervention strategy for endocrine resistance in breast cancer
To Be Published
|
|
4Q3P
 
 | | Crystal structure of Schistosoma mansoni arginase | | Descriptor: | Arginase, GLYCEROL, MANGANESE (II) ION | | Authors: | Hai, Y, Edwards, J.E, Van Zandt, M.C, Hoffmann, K.F, Christianson, D.W. | | Deposit date: | 2014-04-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Arginase, a Potential Drug Target for the Treatment of Schistosomiasis.
Biochemistry, 53, 2014
|
|
4Q3Y
 
 | |
6ALV
 
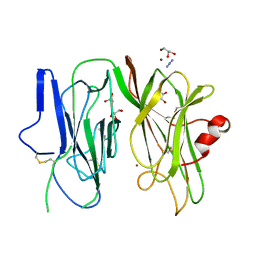 | | Crystal structure of H107A-peptidylglycine alpha-hydroxylating monooxygenase (PHM) mutant (no CuH bound) | | Descriptor: | AZIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-08 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
5L82
 
 | |
8C0L
 
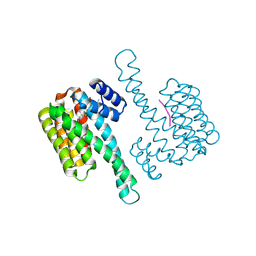 | | FC-THF stabilizer of 14-3-3 and ERalpha | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, Fusicoccin A-THF | | Authors: | Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-17 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Stabilization of the Estrogen receptor alpha - 14-3-3 interaction as a potential intervention strategy for endocrine resistance in breast cancer
To Be Published
|
|
6AFF
 
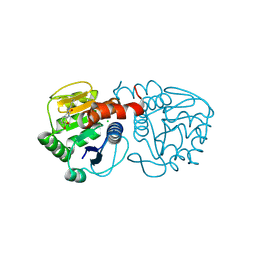 | | DJ-1 with compound 8 | | Descriptor: | CHLORIDE ION, Protein/nucleic acid deglycase DJ-1, methyl 2,3-bis(oxidanylidene)-1~{H}-indole-7-carboxylate | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
4Q3V
 
 | | Crystal structure of Schistosoma mansoni arginase in complex with inhibitor BEC | | Descriptor: | Arginase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Hai, Y, Edwards, J.E, Van Zandt, M.C, Hoffmann, K.F, Christianson, D.W. | | Deposit date: | 2014-04-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Arginase, a Potential Drug Target for the Treatment of Schistosomiasis.
Biochemistry, 53, 2014
|
|
8I0J
 
 | | JB13GH39P28 mutant-D41G | | Descriptor: | CHLORIDE ION, Glycoside hydrolase family 39 beta-xylosidase | | Authors: | Zhou, J.P, Cao, L.J, Lin, M.Y, Zhang, R, Huang, Z.X. | | Deposit date: | 2023-01-11 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | beta-Xylosidase JB13GH39P28 (D41G) showing salt/ethanol/trypsin tolerance and transformation of notoginsenosides
To Be Published
|
|
5LKS
 
 | | Structure-function insights reveal the human ribosome as a cancer target for antibiotics | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Myasnikov, A.G, Natchiar, S.K, Nebout, M, Hazemann, I, Imbert, V, Khatter, H, Peyron, J.-F, Klaholz, B.P. | | Deposit date: | 2016-07-23 | | Release date: | 2017-04-26 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure-function insights reveal the human ribosome as a cancer target for antibiotics.
Nat Commun, 7, 2016
|
|
8I0X
 
 | | Beta-Xylosidase JB13GH39P28 showing salt/ethanol/trypsin tolerance, low-pH/low-Temperature activity, and transformation of notoginsenosides | | Descriptor: | Glycoside hydrolase family 39 beta-xylosidase | | Authors: | Zhou, J.P, Cao, L.J, Lin, M.Y, Zhang, R, Huang, Z.X. | | Deposit date: | 2023-01-11 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | beta-Xylosidase JB13GH39P28(D41G)showing salt/ethanol/trypsin tolerance and transformation of notoginsenosides
To Be Published
|
|
6AFJ
 
 | | DJ-1 with compound 13 | | Descriptor: | CHLORIDE ION, Protein/nucleic acid deglycase DJ-1, butyl 2-[2,3-bis(oxidanylidene)indol-1-yl]ethanoate | | Authors: | Caaveiro, J.M.M, Tashiro, S, Tsumoto, K. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Discovery and Optimization of Inhibitors of the Parkinson's Disease Associated Protein DJ-1.
ACS Chem. Biol., 13, 2018
|
|
8I12
 
 | | InuAMN8 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glycosyl hydrolase family 32 exo-inulinase | | Authors: | Zhou, J.P, Cen, X.L, He, L.M, Zhang, R, Huang, Z.X. | | Deposit date: | 2023-01-12 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Cold-active and NaCl-tolerant exo-inulinase InuAMN8.
To Be Published
|
|
5LFR
 
 | |
