1KLU
 
 | | Crystal structure of HLA-DR1/TPI(23-37) complexed with staphylococcal enterotoxin C3 variant 3B2 (SEC3-3B2) | | Descriptor: | ENTEROTOXIN TYPE C-3, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, DR ALPHA CHAIN, ... | | Authors: | Sundberg, E.J, Sawicki, M.W, Andersen, P.S, Sidney, J, Sette, A, Mariuzza, R.A. | | Deposit date: | 2001-12-12 | | Release date: | 2002-08-02 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Minor structural changes in a mutated human melanoma antigen correspond to dramatically enhanced stimulation of a CD4+ tumor-infiltrating lymphocyte line.
J.Mol.Biol., 319, 2002
|
|
6NY8
 
 | |
5LNR
 
 | | Crystal structure of Arabidopsis thaliana Pdx1-PLP complex | | Descriptor: | GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal 5'-phosphate synthase subunit PDX1.3 | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2017-02-22 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
2AH4
 
 | | guanidinobenzoyl-trypsin acyl-enzyme at 1.13 A resolution | | Descriptor: | 4-carbamimidamidobenzoic acid, CALCIUM ION, SULFATE ION, ... | | Authors: | Radisky, E.S, Lee, J.M, Lu, C.J, Koshland Jr, D.E. | | Deposit date: | 2005-07-26 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Insights into the serine protease mechanism from atomic resolution structures of trypsin reaction intermediates
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
6O0L
 
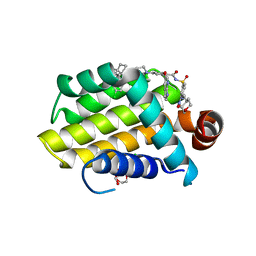 | | crystal structure of BCL-2 G101V mutation with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, CHLORIDE ION, ... | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-16 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
8V6S
 
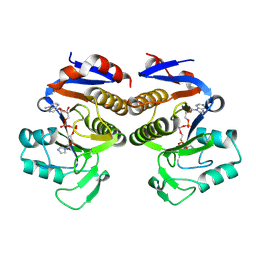 | | Crystal structure of PcThsA in complex with Imidazole Adenine Dinucleotide | | Descriptor: | Thoeris protein ThsA Macro domain-containing protein, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-imidazol-1-yl-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Goulart, C.C, Hartley-Tassell, L, Sorbello, M, Vasquez, E, Mishra, B.P, Holt, S, Gu, W, Kobe, B, Ve, T. | | Deposit date: | 2023-12-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of macro domain-containing Thoeris antiphage defense systems.
Sci Adv, 10, 2024
|
|
6R8S
 
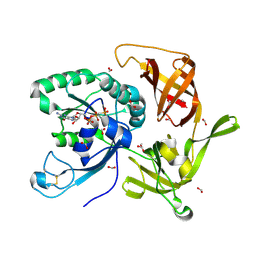 | | Crystal structure of aIF2gamma subunit I181K from archaeon Sulfolobus solfataricus complexed with GDPCP | | Descriptor: | FORMIC ACID, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Kravchenko, O, Arkhipova, V, Gabdulkhakov, A, Stolboushkina, E, Nikonov, O, Garber, M, Nikonov, S. | | Deposit date: | 2019-04-02 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of aIF2gamma subunit I181K from archaeon Sulfolobus solfataricus complexed with GDPCP
To Be Published
|
|
6O0P
 
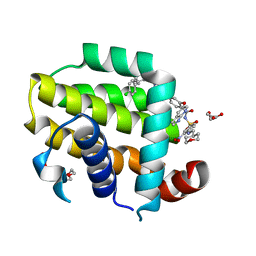 | | crystal structure of BCL-2 G101A mutation with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, DI(HYDROXYETHYL)ETHER | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
1Y1S
 
 | | Crystal Structure of the Uridine Phosphorylase from Salmonella Typhimurium in Complex with Uracil and Sulfate Ion at 2.55A Resolution | | Descriptor: | SULFATE ION, URACIL, Uridine phosphorylase | | Authors: | Gabdoulkhakov, A.G, Dontsova, M.V, Kachalova, G.S, Betzel, C, Ealick, S.E, Mikhailov, A.M. | | Deposit date: | 2004-11-19 | | Release date: | 2005-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structures of Salmonella Typhimurium Uridine Phosphorylase in Native and Three Complexes Forms - with Uridine, Uracil and Sulfate.
To be Published
|
|
2VC5
 
 | | Structural basis for natural lactonase and promiscuous phosphotriesterase activities | | Descriptor: | 1,2-ETHANEDIOL, ARYLDIALKYLPHOSPHATASE, COBALT (II) ION, ... | | Authors: | Elias, M, Dupuy, J, Merone, L, Mandrich, L, Moniot, S, Lecomte, C, Rossi, M, Masson, P, Manco, G, Chabriere, E. | | Deposit date: | 2007-09-18 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Natural Lactonase and Promiscuous Phosphotriesterase Activities.
J.Mol.Biol., 379, 2008
|
|
7LKO
 
 | |
3E1O
 
 | | Crystal structure of E. coli Bacterioferritin (BFR) with two ZN(II) ION sites at the Ferroxidase centre (ZN-BFR). | | Descriptor: | BACTERIOFERRITIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Crow, A, Lawson, T, Lewin, A, Moore, G.R, Le Brun, N. | | Deposit date: | 2008-08-04 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for iron mineralization by bacterioferritin
J.Am.Chem.Soc., 131, 2009
|
|
1Y32
 
 | | NMR structure of humanin in 30% TFE solution | | Descriptor: | Humanin | | Authors: | Benaki, D, Zikos, C, Evangelou, A, Livaniou, E, Vlassi, M, Mikros, E, Pelecanou, M. | | Deposit date: | 2004-11-23 | | Release date: | 2005-02-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of humanin, a peptide against Alzheimer's disease-related neurotoxicity.
Biochem.Biophys.Res.Commun., 329, 2005
|
|
4MCM
 
 | | Human SOD1 C57S Mutant, As-isolated | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Sea, K, Sohn, S.H, Durazo, A, Sheng, Y, Shaw, B, Cao, X, Taylor, A.B, Whitson, L.J, Holloway, S.P, Hart, P.J, Cabelli, D.E, Gralla, E.B, Valentine, J.S. | | Deposit date: | 2013-08-21 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into the role of the unusual disulfide bond in copper-zinc superoxide dismutase.
J.Biol.Chem., 290, 2015
|
|
7LKN
 
 | |
5M1E
 
 | | Crystal structure of N-terminally tagged UbiD from E. coli reconstituted with prFMN cofactor | | Descriptor: | (16~{R})-11,12,14,14-tetramethyl-3,5-bis(oxidanylidene)-8-[(2~{S},3~{S},4~{R})-2,3,4-tris(oxidanyl)-5-phosphonooxy-pentyl]-1,4,6,8-tetrazatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-2(7),9(17),10,12-tetraene-16-sulfonic acid, 3-octaprenyl-4-hydroxybenzoate carboxy-lyase, MANGANESE (II) ION, ... | | Authors: | Marshall, S.A, Leys, D. | | Deposit date: | 2016-10-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Oxidative Maturation and Structural Characterization of Prenylated FMN Binding by UbiD, a Decarboxylase Involved in Bacterial Ubiquinone Biosynthesis.
J. Biol. Chem., 292, 2017
|
|
4QPG
 
 | | Crystal structure of empty hepatitis A virus | | Descriptor: | CHLORIDE ION, Capsid protein VP0, Capsid protein VP1, ... | | Authors: | Wang, X, Ren, J, Gao, Q, Hu, Z, Sun, Y, Li, X, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Hepatitis A virus and the origins of picornaviruses.
Nature, 517, 2015
|
|
2VAD
 
 | | Monomeric red fluorescent protein, DsRed.M1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, RED FLUORESCENT PROTEIN, ... | | Authors: | Strongin, D.E, Bevis, B, Khuong, N, Downing, M.E, Strack, R.L, Sundaram, K, Glick, B.S, Keenan, R.J. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Rearrangements Near the Chromophore Influence the Maturation Speed and Brightness of Dsred Variants.
Protein Eng.Des.Sel., 20, 2007
|
|
5LSP
 
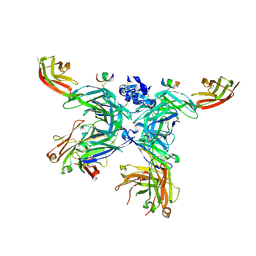 | | 107_A07 Fab in complex with fragment of the Met receptor | | Descriptor: | 107_A07 Fab heavy chain, 107_A07 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | DiCara, D, Chirgadze, D.Y, Pope, A, Karatt-Vellatt, A, Winter, A, van den Heuvel, J, Gherardi, E, McCafferty, J. | | Deposit date: | 2016-09-05 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Characterization and structural determination of a new anti-MET function-blocking antibody with binding epitope distinct from the ligand binding domain.
Sci Rep, 7, 2017
|
|
6REG
 
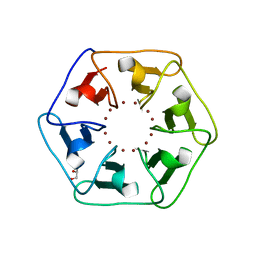 | | Crystal structure of Pizza6-S with Zn2+ | | Descriptor: | GLYCEROL, Pizza6-S, ZINC ION | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
6MHD
 
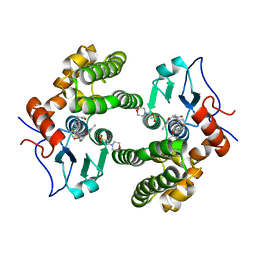 | | Glutathione S-Transferase Omega 1 bound to covalent inhibitor 44 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETONE, Glutathione S-transferase omega-1, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure-Based Design of N-(5-Phenylthiazol-2-yl)acrylamides as Novel and Potent Glutathione S-Transferase Omega 1 Inhibitors.
J. Med. Chem., 62, 2019
|
|
6REK
 
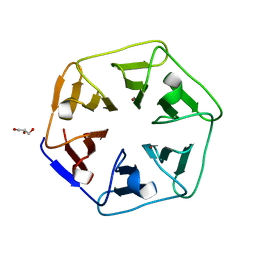 | | Crystal structure of Pizza6-SH with Cu2+ | | Descriptor: | COPPER (II) ION, GLYCEROL, Pizza6-SH | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
7LKM
 
 | |
5LTT
 
 | |
5UN1
 
 | | Crystal structure of GluN1/GluN2B delta-ATD NMDA receptor | | Descriptor: | (5S,10R)-5-methyl-10,11-dihydro-5H-5,10-epiminodibenzo[a,d][7]annulene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Song, X, Gouaux, E. | | Deposit date: | 2017-01-30 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Mechanism of NMDA receptor channel block by MK-801 and memantine.
Nature, 556, 2018
|
|
