6VLF
 
 | | Crystal structure of mouse alpha 1,6-fucosyltransferase, FUT8 in its Apo-form | | Descriptor: | 1,2-ETHANEDIOL, Alpha-(1,6)-fucosyltransferase, SULFATE ION | | Authors: | Jarva, M.A, Dramicanin, M, Lingford, J.P, Mao, R, John, A, Goddard-Borger, E. | | Deposit date: | 2020-01-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of substrate recognition and catalysis by fucosyltransferase 8.
J.Biol.Chem., 295, 2020
|
|
6XVK
 
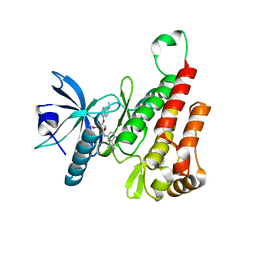 | | Crystal structure of the KDR (VEGFR2) kinase domain in complex with a type-II inhibitor bearing an acrylamide | | Descriptor: | Vascular endothelial growth factor receptor 2, ~{N}-(4,4-dimethyl-2-prop-1-ynyl-3,1-benzoxazin-6-yl)-2-[3-methoxy-5-(7-methoxyquinolin-4-yl)oxy-pyridin-2-yl]ethanamide | | Authors: | Schimpl, M, McAuley, K, Hoyt, E.A, Thomas, M, Bodnarchuk, M.S, Lewis, H.J, Barratt, D, Deery, M.J, Ogg, D.J, Bernardes, G.J.L, Ward, R.A, Kettle, J.G, Waring, M.J. | | Deposit date: | 2020-01-22 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Alkynyl Benzoxazines and Dihydroquinazolines as Cysteine Targeting Covalent Warheads and Their Application in Identification of Selective Irreversible Kinase Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6VKK
 
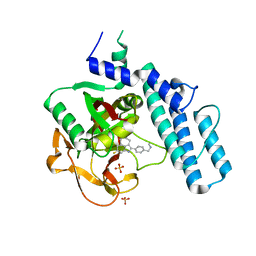 | |
7ZTR
 
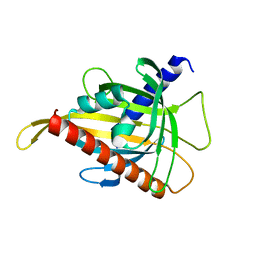 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform, W232F mutant | | Descriptor: | Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Silkworm carotenoprotein as an efficient carotenoid extractor, solubilizer and transporter.
Int.J.Biol.Macromol., 223, 2022
|
|
7ZVR
 
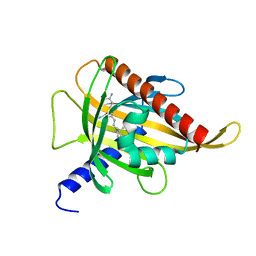 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) complexed with zeaxanthin | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the carotenoid binding and transport function of a START domain.
Structure, 30, 2022
|
|
7ZVQ
 
 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) in the apoform, S206V mutant | | Descriptor: | Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the carotenoid binding and transport function of a START domain.
Structure, 30, 2022
|
|
6XV9
 
 | | Crystal structure of the kinase domain of human c-KIT in complex with a type-II inhibitor | | Descriptor: | Mast/stem cell growth factor receptor Kit,Mast/stem cell growth factor receptor Kit, ~{N}-[3-[(dimethylamino)methyl]-5-methyl-phenyl]-2-[3-methoxy-5-(7-methoxyquinolin-4-yl)oxy-pyridin-2-yl]ethanamide | | Authors: | Ogg, D.J, Howard, T, McAuley, K, Hoyt, E.A, Thomas, M, Bodnarchuk, M.S, Lewis, H.J, Barratt, D, Deery, M.J, Bernardes, G.J.L, Ward, R.A, Kettle, J.G, Waring, M.J. | | Deposit date: | 2020-01-21 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Alkynyl Benzoxazines and Dihydroquinazolines as Cysteine Targeting Covalent Warheads and Their Application in Identification of Selective Irreversible Kinase Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6VLD
 
 | | Crystal structure of human alpha 1,6-fucosyltransferase, FUT8 bound to GDP and A2SGP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASPARAGINE, Alpha-(1,6)-fucosyltransferase, ... | | Authors: | Jarva, M.A, Dramicanin, M, Lingford, J.P, Mao, R, John, A, Goddard-Borger, E.D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis of substrate recognition and catalysis by fucosyltransferase 8.
J.Biol.Chem., 295, 2020
|
|
6BXQ
 
 | | Crystal Structure of HLA-B*57:01 with an HIV peptide RKV | | Descriptor: | Beta-2-microglobulin, HIV peptide RKV, MHC class I antigen | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-12-18 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification of Native and Posttranslationally Modified HLA-B*57:01-Restricted HIV Envelope Derived Epitopes Using Immunoproteomics.
Proteomics, 18, 2018
|
|
6C5T
 
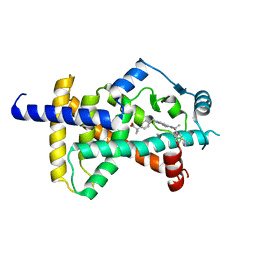 | | PPARg LBD bound to SR11023 | | Descriptor: | 2-{4-[(5-{[(1R)-1-(3-cyclopropylphenyl)ethyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]phenyl}-2-methylpropanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Bruning, J.B, Frkic, R.L, Griffin, P.R. | | Deposit date: | 2018-01-16 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | PPAR gamma in Complex with an Antagonist and Inverse Agonist: a Tumble and Trap Mechanism of the Activation Helix.
iScience, 5, 2018
|
|
6Z1Y
 
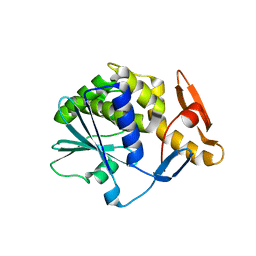 | | Crystal structure of type-I ribosome-inactivating protein trichobakin (TBK) | | Descriptor: | SODIUM ION, Trichobakin | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Britikov, V.V, Bocharov, E.V, Britikova, E.V, Le, T.B.T, Phan, C.V, Popov, V.O, Usanov, S.A. | | Deposit date: | 2020-05-14 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of type-I ribosome-inactivating protein trichobakin (TBK)
To Be Published
|
|
6Z6A
 
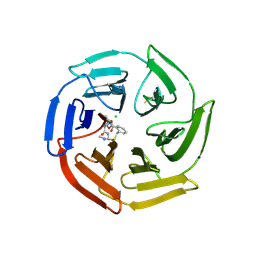 | | Keap1 macrocycle complex | | Descriptor: | (5S,8R)-8-[[(2S)-1-ethanoylpyrrolidin-2-yl]carbonylamino]-N,N-dimethyl-7,11-bis(oxidanylidene)-10-oxa-3-thia-6-azabicyclo[10.4.0]hexadeca-1(16),12,14-triene-5-carboxamide, CHLORIDE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Johansson, P. | | Deposit date: | 2020-05-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mining Natural Products for Macrocycles to Drug Difficult Targets.
J.Med.Chem., 64, 2021
|
|
5U5I
 
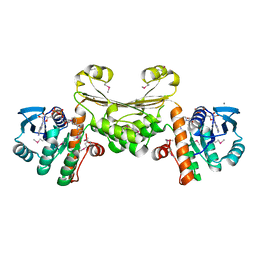 | |
6VLE
 
 | | Crystal structure of human alpha 1,6-fucosyltransferase, FUT8 in its Apo-form | | Descriptor: | Alpha-(1,6)-fucosyltransferase, SULFATE ION | | Authors: | Jarva, M.A, Dramicanin, M, Lingford, J.P, Mao, R, John, A, Goddard-Borger, E.D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis of substrate recognition and catalysis by fucosyltransferase 8.
J.Biol.Chem., 295, 2020
|
|
6XM1
 
 | |
6XU4
 
 | | Crystal structure of the genetically-encoded FGCaMP calcium indicator in its calcium-bound state | | Descriptor: | CALCIUM ION, FGCamp | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Barykina, N.V, Subach, O.M, Subach, F.V. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | FGCaMP7, an Improved Version of Fungi-Based Ratiometric Calcium Indicator for In Vivo Visualization of Neuronal Activity.
Int J Mol Sci, 21, 2020
|
|
6XMD
 
 | |
6W9D
 
 | |
6HC0
 
 | |
6HVD
 
 | | Human SLK bound to a maleimide inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-(1,3-benzothiazol-2-ylamino)-4-(2-methoxyphenyl)pyrrole-2,5-dione, STE20-like serine/threonine-protein kinase, ... | | Authors: | Sorrell, F.J, Berger, B.T, Salah, E, Serafim, R.A.M, Savitsky, P.A, Krojer, T, Bailey, H.J, Pinkas, D, Burgess-Brown, N.A, von Delft, F, Knapp, S, Arrowsmith, C, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2018-10-10 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Human SLK bound to a maleimide inhibitor
To Be Published
|
|
6HG9
 
 | |
6VZH
 
 | | Structure of Human Vaccinia-related Kinase 1 (VRK1) Bound to LDSM311 | | Descriptor: | (7~{R})-2-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]amino]-5,7-dimethyl-8-prop-2-ynyl-7~{H}-pteridin-6-one, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | dos Reis, C.V, Dutra, L.A, Gama, F, Ferreira, M, Mascarello, A, Azevedo, H, Guimaraes, C, Massirer, K.B, Arruda, P, Edwards, A.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-28 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of Human Vaccinia-related Kinase 1 (VRK1) Bound to LDSM311
To be Published
|
|
8AQ6
 
 | |
8AQI
 
 | |
6WAQ
 
 | |
