3UXF
 
 | |
4O91
 
 | | Crystal Structure of type II inhibitor NG25 bound to TAK1-TAB1 | | Descriptor: | Mitogen-activated protein kinase kinase kinase 7/TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera, N-{4-[(4-ethylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-4-methyl-3-(1H-pyrrolo[2,3-b]pyridin-4-yloxy)benzamide | | Authors: | Gurbani, D, Hunter, J.C, Tan, L, Westover, K.D. | | Deposit date: | 2013-12-31 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Discovery of Type II Inhibitors of TGF beta-Activated Kinase 1 (TAK1) and Mitogen-Activated Protein Kinase Kinase Kinase Kinase 2 (MAP4K2).
J.Med.Chem., 58, 2015
|
|
3UM0
 
 | |
3UME
 
 | | Structure of pB intermediate of Photoactive yellow protein (PYP) at pH 7 | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Tripathi, S, Srajer, V, Purwar, N, Henning, R, Schmidt, M. | | Deposit date: | 2011-11-13 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | pH Dependence of the Photoactive Yellow Protein Photocycle Investigated by Time-Resolved Crystallography.
Biophys.J., 102, 2012
|
|
4OV5
 
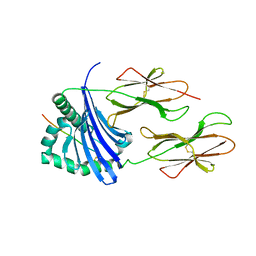 | | Structure of HLA-DR1 with a bound peptide with non-optimal alanine in the P1 pocket | | Descriptor: | HLA class I histocompatibility antigen, A-2 alpha chain, HLA class II histocompatibility antigen, ... | | Authors: | Trenh, P, Yin, L, Stern, L.J. | | Deposit date: | 2014-02-20 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Susceptibility to HLA-DM Protein Is Determined by a Dynamic Conformation of Major Histocompatibility Complex Class II Molecule Bound with Peptide.
J.Biol.Chem., 289, 2014
|
|
3UYN
 
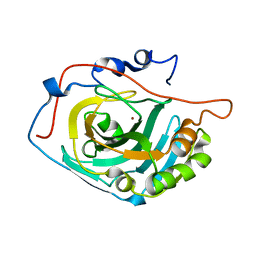 | | HCA 3 | | Descriptor: | Carbonic anhydrase 3, ZINC ION | | Authors: | Fisher, Z, McKenna, R. | | Deposit date: | 2011-12-06 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and kinetic analysis of proton shuttle residues in the active site of human carbonic anhydrase III.
Proteins, 68, 2007
|
|
2RGV
 
 | |
4O97
 
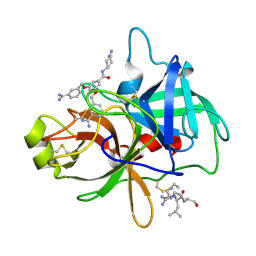 | | Crystal structure of matriptase in complex with inhibitor | | Descriptor: | N-(trans-4-aminocyclohexyl)-3,5-bis[(3-carbamimidoylbenzyl)oxy]benzamide, Peptide CGLR, Suppressor of tumorigenicity 14 protein | | Authors: | Rao, K.N, Chandra, B.R, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S, Subramanya, H.S. | | Deposit date: | 2014-01-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-guided discovery of 1,3,5 tri-substituted benzenes as potent and selective matriptase inhibitors exhibiting in vivo antitumor efficacy.
Bioorg.Med.Chem., 22, 2014
|
|
3UMH
 
 | | X-ray structure of the E2 domain of the human amyloid precursor protein (APP) in complex with cadmium | | Descriptor: | ACETATE ION, Amyloid beta A4 protein, CADMIUM ION | | Authors: | Dahms, S.O, Konnig, I, Roeser, D, Guhrs, K.H, Than, M.E. | | Deposit date: | 2011-11-13 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal Binding Dictates Conformation and Function of the Amyloid Precursor Protein (APP) E2 Domain.
J.Mol.Biol., 416, 2012
|
|
4OW3
 
 | | Thermolysin structure determined by free-electron laser | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Hattne, J, Echols, N, Tran, R, Kern, J, Gildea, R.J, Brewster, A.S, Alonso-Mori, R, Glockner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, White, W.E, Schafer, D.W, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Glatzel, P, Zwart, P.H, Grosse-Kunstleve, R.W, Bogan, M.J, Messerschmidt, M, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Yano, J, Bergmann, U, Yachandra, V.K, Adams, P.D, Sauter, N.K. | | Deposit date: | 2014-01-30 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Accurate macromolecular structures using minimal measurements from X-ray free-electron lasers.
Nat.Methods, 11, 2014
|
|
3UYW
 
 | | Crystal structures of globular head of 2009 pandemic H1N1 hemagglutinin | | Descriptor: | 2-AMINOETHANESULFONIC ACID, Hemagglutinin | | Authors: | Xuan, C.L, Shi, Y, Qi, J.X, Xiao, H.X, Gao, G.F. | | Deposit date: | 2011-12-06 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Structural vaccinology: structure-based design of influenza A virus hemagglutinin subtype-specific subunit vaccines
Protein Cell, 2, 2011
|
|
4O9G
 
 | | Crystal structure of the H51N mutant of the 3,4-ketoisomerase QdtA from Thermoanaerobacterium thermosaccharolyticum in complex with TDP-4-keto-6-deoxyglucose | | Descriptor: | (2S)-1-[3-[(2S)-2-oxidanylpropoxy]-2-[[(2S)-2-oxidanylpropoxy]methyl]-2-[[(2R)-2-oxidanylpropoxy]methyl]propoxy]propan-2-ol, 1,2-ETHANEDIOL, QdtA, ... | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2014-01-02 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The molecular architecture of QdtA, a sugar 3,4-ketoisomerase from Thermoanaerobacterium thermosaccharolyticum.
Protein Sci., 23, 2014
|
|
4OWE
 
 | | PtCl6 binding to HEWL | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Starkey, V.L, Lamplough, L, Kaenket, S, Helliwell, J.R. | | Deposit date: | 2014-01-31 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | The binding of platinum hexahalides (Cl, Br and I) to hen egg-white lysozyme and the chemical transformation of the PtI6 octahedral complex to a PtI3 moiety bound to His15.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4O9S
 
 | | Crystal structure of Retinol-Binding Protein 4 (RBP4)in complex with a non-retinoid ligand | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(12),2(6),8,10-tetraen-12-yl)piperazin-1-yl]-2-[2-(trifluoromethyl)phenyl]ethanone, CHLORIDE ION, ... | | Authors: | Wang, Z, Johnstone, S, Walker, N.P. | | Deposit date: | 2014-01-02 | | Release date: | 2014-07-02 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-assisted discovery of the first non-retinoid ligands for Retinol-Binding Protein 4.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3V01
 
 | | Discovery of Novel Allosteric MEK Inhibitors Possessing Classical and Non-classical Bidentate Ser212 Interactions. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Heald, R, Jackson, P, Savy, P, Jones, M, Gancia, E, Burton, B, Newman, R, Boggs, J, Chan, E, Chan, J, Choo, E, Merchant, M, Ultsch, M, Wiesmann, C, Belvin, M, Price, S. | | Deposit date: | 2011-12-07 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Discovery of Novel Allosteric Mitogen-Activated Protein Kinase Kinase (MEK) 1,2 Inhibitors Possessing Bidentate Ser212 Interactions.
J.Med.Chem., 55, 2012
|
|
3UNG
 
 | | Structure of the Cmr2 subunit of the CRISPR RNA silencing complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Cmr2dHD, ... | | Authors: | Cocozaki, A.I, Ramia, N.F, Shao, Y, Hale, C.R, Terns, R.M, Terns, M.P, Li, H. | | Deposit date: | 2011-11-15 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of the Cmr2 Subunit of the CRISPR-Cas RNA Silencing Complex.
Structure, 20, 2012
|
|
4OWN
 
 | | Anthranilate phosphoribosyl transferase from Mycobacterium tuberculosis in complex with 5-fluoroanthranilate, PRPP and Magnesium | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-amino-5-fluorobenzoic acid, Anthranilate phosphoribosyltransferase, ... | | Authors: | Castell, A, Cookson, T.V.M, Short, F.L, Lott, J.S. | | Deposit date: | 2014-02-02 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Alternative substrates reveal catalytic cycle and key binding events in the reaction catalysed by anthranilate phosphoribosyltransferase from Mycobacterium tuberculosis.
Biochem.J., 461, 2014
|
|
4OX3
 
 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | Descriptor: | PHOSPHATE ION, Putative carboxypeptidase YodJ, ZINC ION | | Authors: | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-02-04 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
3UNP
 
 | | Structure of human SUN2 SUN domain | | Descriptor: | ACETYL GROUP, SUN domain-containing protein 2 | | Authors: | Zhou, Z.C, Greene, M.I. | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of Sad1-UNC84 homology (SUN) domain defines features of molecular bridge in nuclear envelope
J.Biol.Chem., 287, 2012
|
|
3V2Z
 
 | | Nitrite Bound Chlorin Substituted Myoglobin- Method 2 | | Descriptor: | Fe(III) pyropheophorbide-a methyl ester, Myoglobin, NITRITE ION, ... | | Authors: | Yi, J, Thomas, L.M, Ricther-Addo, G.B. | | Deposit date: | 2011-12-12 | | Release date: | 2012-11-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Distal pocket control of nitrite binding in myoglobin
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4OXF
 
 | |
3UNT
 
 | | tRNA-guanine transglycosylase E339Q mutant | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Jakobi, S, Heine, A, Klebe, G. | | Deposit date: | 2011-11-16 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Hot-spot analysis to dissect the functional protein-protein interface of a tRNA-modifying enzyme.
Proteins, 82, 2014
|
|
3V36
 
 | | Aldose reductase complexed with glceraldehyde | | Descriptor: | Aldose reductase, D-Glyceraldehyde, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zheng, X, Zhang, L, Chen, Y, Luo, H, Hu, X. | | Deposit date: | 2011-12-13 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Partial inhibition of aldose reductase by nitazoxanide and its molecular basis.
Chemmedchem, 7, 2012
|
|
4OY2
 
 | | Crystal structure of TAF1-TAF7, a TFIID subcomplex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 7, ... | | Authors: | Bhattacharya, S, Lou, X, Rajashankar, K, Jacobson, R.H, Webb, P. | | Deposit date: | 2014-02-10 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insight into TAF1-TAF7, a subcomplex of transcription factor II D.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3UO1
 
 | | Structure of a monoclonal antibody complexed with its MHC-I antigen | | Descriptor: | ANTI-MHC-I MONOCLONAL ANTIBODY, 64-3-7 H CHAIN, 64-3-7 L CHAIN, ... | | Authors: | Margulies, D.H, Mage, M.G, Wang, R, Natarajan, K. | | Deposit date: | 2011-11-16 | | Release date: | 2012-07-25 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | The Peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics.
J.Immunol., 189, 2012
|
|
