3USY
 
 | |
4ORX
 
 | | Three-dimensional structure of the C65A-K59A double mutant of Human lipocalin-type Prostaglandin D Synthase holo-form | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, Prostaglandin-H2 D-isomerase, SULFATE ION | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3UQV
 
 | | Bovine trypsin variant X(triplePhe227) in complex with small molecule inhibitor | | Descriptor: | 3-(3-carbamimidoylphenyl)-N-(2'-sulfamoylbiphenyl-4-yl)-1,2-oxazole-4-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-11-21 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
4OS7
 
 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK607 (bicyclic) | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-09-24 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
3UT7
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
3UR2
 
 | | Crystal Structure of PTE mutant H254G/H257W/L303T/K185R/I274N/A80V | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, IMIDAZOLE, ... | | Authors: | Tsai, P, Fox, N.G, Li, Y, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents.
Biochemistry, 51, 2012
|
|
4OGE
 
 | | Crystal structure of the Type II-C Cas9 enzyme from Actinomyces naeslundii | | Descriptor: | HNH endonuclease domain protein, MAGNESIUM ION, SPERMIDINE, ... | | Authors: | Jiang, F, Ma, E, Lin, S, Doudna, J.A. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structures of Cas9 endonucleases reveal RNA-mediated conformational activation.
Science, 343, 2014
|
|
4OSL
 
 | | Crystal structure of TAL effector reveals the recognition between histidine and guanine | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*CP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*GP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3, ... | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
3URC
 
 | | T181G mutant of alpha-Lytic Protease | | Descriptor: | Alpha-lytic protease, GLYCEROL, SULFATE ION | | Authors: | Kelch, B.A, Agard, D.A. | | Deposit date: | 2011-11-22 | | Release date: | 2012-05-23 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Functional modulation of a protein folding landscape via side-chain distortion.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4OT3
 
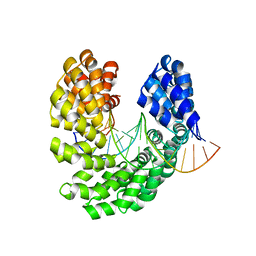 | | Crystal structure of the S505L mutant of TAL effector dHax3 | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*AP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3 | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
4OHM
 
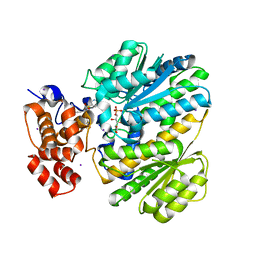 | | Human GKRP bound to AMG-0771 and sorbitol-6-phosphate | | Descriptor: | (2S)-2-{4-[(2S)-4-[(6-aminopyridin-3-yl)sulfonyl]-2-(prop-1-yn-1-yl)piperazin-1-yl]phenyl}-3,3,3-trifluoropropane-1,2-diol, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | Jordan, S.R, Chmait, S. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Small molecule disruptors of the glucokinase-glucokinase regulatory protein interaction: 3. Structure-activity relationships within the aryl carbinol region of the N-arylsulfonamido-N'-arylpiperazine series.
J.Med.Chem., 57, 2014
|
|
3URM
 
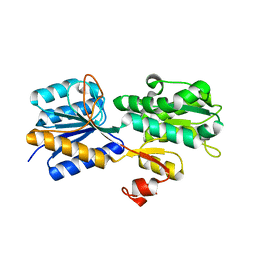 | | Crystal structure of the periplasmic sugar binding protein ChvE | | Descriptor: | Multiple sugar-binding periplasmic receptor ChvE, beta-D-galactopyranose | | Authors: | Hu, X, Zhao, J, Binns, A, Degrado, W. | | Deposit date: | 2011-11-22 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3USD
 
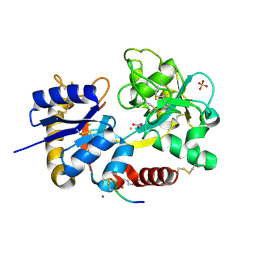 | | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Imidazol (1,2 a) pyridine3-yl-acitic acid at 2.4 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-terminal peptide of Lactotransferrin, ... | | Authors: | Shukla, P.K, Gautam, L, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Imidazol (1,2 a) pyridine3-yl-acitic acid at 2.4 A Resolution
To be Published
|
|
2R60
 
 | |
3UUG
 
 | | Crystal structure of the periplasmic sugar binding protein ChvE | | Descriptor: | Multiple sugar-binding periplasmic receptor ChvE, beta-D-glucopyranuronic acid | | Authors: | Hu, X, Zhao, J, Binns, A, Degrado, W. | | Deposit date: | 2011-11-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4OI5
 
 | | Glycerol-free structure of thermolysin in complex with ubtln58 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, P-((((benzyloxy)carbonyl)amino)methyl)-N-((S)-1-((3,3-dimethylbutyl)amino)-4-methyl-1-oxopentan-2-yl)phosphonamidic acid, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2014-01-18 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Methyl, Ethyl, Propyl, Butyl: Futile But Not for Water, as the Correlation of Structure and Thermodynamic Signature Shows in a Congeneric Series of Thermolysin Inhibitors.
Chemmedchem, 9, 2014
|
|
3USL
 
 | | Crystal Structure of LeuT bound to L-selenomethionine in space group C2 from lipid bicelles | | Descriptor: | ACETATE ION, IODIDE ION, PHOSPHOCHOLINE, ... | | Authors: | Wang, H, Elferich, J, Gouaux, E. | | Deposit date: | 2011-11-23 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4OVI
 
 | | Phenylacetone monooxygenase: oxidised enzyme in complex with APADP | | Descriptor: | 3-ACETYLPYRIDINE ADENINE DINUCLEOTIDE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Martinoli, C, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2013-09-19 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Beyond the Protein Matrix: Probing Cofactor Variants in a Baeyer-Villiger Oxygenation Reaction.
Acs Catalysis, 3, 2013
|
|
4OIJ
 
 | | X-ray crystal structure of racemic non-glycosylated chemokine Ser-CCL1 | | Descriptor: | C-C motif chemokine 1, D-Ser-CCL1, SULFATE ION | | Authors: | Okamoto, R, Mandal, K, Sawaya, M.R, Kajihara, Y, Yeates, T.O, Kent, S.B.H. | | Deposit date: | 2014-01-19 | | Release date: | 2014-05-07 | | Last modified: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | (Quasi-)Racemic X-ray Structures of Glycosylated and Non-Glycosylated Forms of the Chemokine Ser-CCL1 Prepared by Total Chemical Synthesis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3USU
 
 | | Crystal structure of Butea monosperma seed lectin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Abhilash, J, Geethanandan, K, Bharath, S.R, Sadasivan, C, Haridas, M. | | Deposit date: | 2011-11-24 | | Release date: | 2012-01-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure of Butea monosperma seed lectin
To be Published
|
|
4OW0
 
 | |
3UWD
 
 | | Crystal Structure of Phosphoglycerate Kinase from Bacillus Anthracis | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Zheng, H, Chruszcz, M, Porebski, P, Kudritska, M, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-01 | | Release date: | 2012-01-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structures of putative phosphoglycerate kinases from B. anthracis and C. jejuni.
J.Struct.Funct.Genom., 13, 2012
|
|
3UTX
 
 | |
4O6P
 
 | | Structural and functional studies the characterization of C58G/C70G mutant in Cys4 Zinc-finger motif in the recombination mediator protein RecR | | Descriptor: | Recombination protein RecR, ZINC ION | | Authors: | Tang, Q, Liu, Y.P, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-12-23 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional characterization of Cys4 zinc finger motif in the recombination mediator protein RecR.
DNA Repair (Amst.), 24, 2014
|
|
3UU4
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-21' mutant reduced in the crystal in a locally-closed conformation (LC1 subtype) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
