3HZO
 
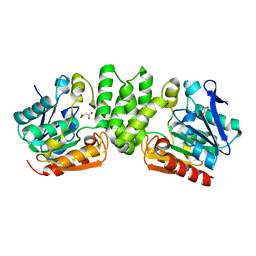 | |
3I00
 
 | | Crystal Structure of the huntingtin interacting protein 1 coiled coil domain | | Descriptor: | Huntingtin-interacting protein 1 | | Authors: | Wilbur, J.D, Hwang, P.K, Brodsky, F.M, Fletterick, R.J. | | Deposit date: | 2009-06-24 | | Release date: | 2010-02-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Accommodation of structural rearrangements in the huntingtin-interacting protein 1 coiled-coil domain.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3I0C
 
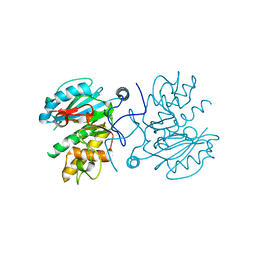 | | Crystal structure of GTB C80S/C196S unliganded | | Descriptor: | ABO glycosyltransferase | | Authors: | Schuman, B, Persson, M, Landry, R.C, Polakowski, R, Weadge, J.T, Seto, N.O.L, Borisova, S, Palcic, M.M, Evans, S.V. | | Deposit date: | 2009-06-25 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Cysteine-to-serine mutants dramatically reorder the active site of human ABO(H) blood group B glycosyltransferase without affecting activity: structural insights into cooperative substrate binding
J.Mol.Biol., 402, 2010
|
|
3HPJ
 
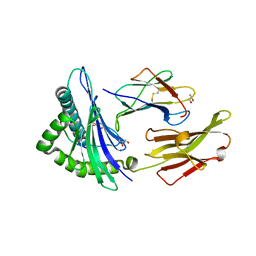 | |
3I0H
 
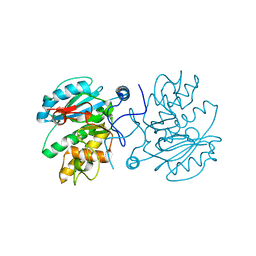 | | Crystal structure of GTB C80S/C196S/C209S unliganded | | Descriptor: | ABO glycosyltransferase | | Authors: | Schuman, B, Persson, M, Landry, R.C, Polakowski, R, Weadge, J.T, Seto, N.O.L, Borisova, S, Palcic, M.M, Evans, S.V. | | Deposit date: | 2009-06-25 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cysteine-to-serine mutants dramatically reorder the active site of human ABO(H) blood group B glycosyltransferase without affecting activity: structural insights into cooperative substrate binding
J.Mol.Biol., 402, 2010
|
|
3HQZ
 
 | | Discovery of novel inhibitors of PDE10A | | Descriptor: | 2-{[4-(4-pyridin-4-yl-1H-pyrazol-3-yl)phenoxy]methyl}quinoline, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Pandit, J, Marr, E.S. | | Deposit date: | 2009-06-08 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of a Novel Class of Phosphodiesterase 10A Inhibitors and Identification of Clinical Candidate 2-[4-(1-Methyl-4-pyridin-4-yl-1H-pyrazol-3-yl)-phenoxymethyl]-quinoline (PF-2545920) for the Treatment of Schizophrenia
J.Med.Chem., 52, 2009
|
|
3HRG
 
 | |
3HRO
 
 | | Crystal structure of a C-terminal coiled coil domain of Transient receptor potential (TRP) channel subfamily P member 2 (TRPP2, polycystic kidney disease 2) | | Descriptor: | Transient receptor potential (TRP) channel subfamily P member 2 (TRPP2), also called Polycystin-2 or polycystic kidney disease 2(PKD2) | | Authors: | Yu, Y, Ulbrich, M.H, Li, M.-H, Buraei, Z, Chen, X.-Z, Ong, A.C.M, Tong, L, Isacoff, E.Y, Yang, J. | | Deposit date: | 2009-06-09 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and molecular basis of the assembly of the TRPP2/PKD1 complex.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3I3C
 
 | | Crystal Structural of CBX5 Chromo Shadow Domain | | Descriptor: | Chromobox protein homolog 5, SODIUM ION | | Authors: | Amaya, M.F, Li, Z, Li, Y, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structural of CBX5 Chromo Shadow Domain
To be Published
|
|
3HRT
 
 | | Crystal Structure of ScaR with bound Cd2+ | | Descriptor: | CADMIUM ION, Metalloregulator ScaR, SULFATE ION | | Authors: | Stoll, K.E, Draper, W.E, Kliegman, J.I, Golynskiy, M.V, Brew-Appiah, R.A.T, Brown, H.K, Breyer, W.A, Jakubovics, N.S, Jenkinson, H.F, Brennan, R.B, Cohen, S.M, Glasfeld, A. | | Deposit date: | 2009-06-09 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization and structure of the manganese-responsive transcriptional regulator ScaR.
Biochemistry, 48, 2009
|
|
3HSE
 
 | | Crystal structure of Staphylococcus aureus protein SarZ in reduced form | | Descriptor: | HTH-type transcriptional regulator sarZ | | Authors: | Poor, C.B, Duguid, E, Rice, P.A, He, C. | | Deposit date: | 2009-06-10 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus.
J.Biol.Chem., 284, 2009
|
|
3HSM
 
 | | Crystal structure of distal N-terminal beta-trefoil domain of Ryanodine Receptor type 1 | | Descriptor: | Ryanodine receptor 1 | | Authors: | Amador, F.J, Liu, S, Ishiyama, N, Plevin, M.J, Wilson, A, MacLennan, D.H, Ikura, M. | | Deposit date: | 2009-06-10 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of type I ryanodine receptor amino-terminal beta-trefoil domain reveals a disease-associated mutation "hot spot" loop
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HSR
 
 | | Crystal structure of Staphylococcus aureus protein SarZ in mixed disulfide form | | Descriptor: | ACETATE ION, GLYCEROL, HTH-type transcriptional regulator sarZ, ... | | Authors: | Poor, C.B, Duguid, E, Rice, P.A, He, C. | | Deposit date: | 2009-06-10 | | Release date: | 2009-07-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus.
J.Biol.Chem., 284, 2009
|
|
3HSZ
 
 | | Crystal structure of E. coli HPPK(F123A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-06-11 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Pterin-binding site mutation Y53A, N55A or F123A and activity of E. coli HPPK
To be Published
|
|
3HT5
 
 | |
3HU8
 
 | | 2-ethoxyphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 2-ethoxyphenol, Lysozyme, PHOSPHATE ION | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3I6J
 
 | | Ribonuclease A by Classical hanging drop method after high X-Ray dose on ESRF ID14-2 beamline | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Pechkova, E, Tripathi, S.K, Ravelli, R, McSweeney, S, Nicolini, C. | | Deposit date: | 2009-07-07 | | Release date: | 2010-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic structure and radiation resistance of langmuir-blodgett protein crystals
To be Published
|
|
3I6S
 
 | | Crystal Structure of the plant subtilisin-like protease SBT3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Subtilisin-like protease, alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rose, R, Ottmann, C. | | Deposit date: | 2009-07-07 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for Ca2+-independence and activation by homodimerization of tomato subtilase 3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HMH
 
 | | Crystal structure of the second bromodomain of human TBP-associated factor RNA polymerase 1-like (TAF1L) | | Descriptor: | Transcription initiation factor TFIID 210 kDa subunit | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Zhang, Y, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-29 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3HMV
 
 | | Catalytic domain of human phosphodiesterase 4B2B in complex with a tetrahydrobenzothiophene inhibitor | | Descriptor: | (6S)-6-methyl-2-{[(2-nitrophenyl)carbonyl]amino}-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Somers, D.O, Neu, M. | | Deposit date: | 2009-05-29 | | Release date: | 2010-06-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Identification of PDE4B Over 4D subtype-selective inhibitors revealing an unprecedented binding mode
Bioorg.Med.Chem., 17, 2009
|
|
3HND
 
 | | Crystal structure of human ribonucleotide reductase 1 bound to the effector TTP and substrate GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Fairman, J.W, Wijerathna, S.R, Xu, H, Dealwis, C.G. | | Deposit date: | 2009-05-31 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3HOB
 
 | |
3HP6
 
 | |
3HNZ
 
 | |
3HQ4
 
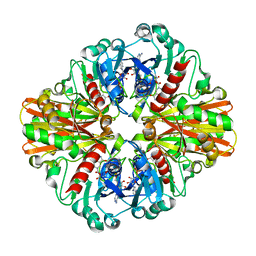 | | Crystal Structure of C151S mutant of Glyceraldehyde-3-phosphate dehydrogenase 1 (GAPDH1) complexed with NAD from Staphylococcus aureus MRSA252 at 2.2 angstrom resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2009-06-05 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
