5U8L
 
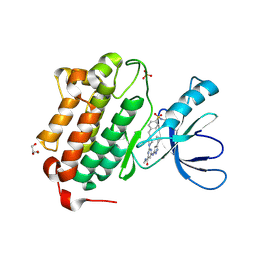 | | Crystal structure of EGFR kinase domain in complex with a sulfonyl fluoride probe XO44 | | Descriptor: | 4-[(4-{4-[(3-cyclopropyl-1H-pyrazol-5-yl)amino]-6-[(prop-2-yn-1-yl)carbamoyl]pyrimidin-2-yl}piperazin-1-yl)methyl]benzene-1-sulfonyl fluoride, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S, Ferre, R. | | Deposit date: | 2016-12-14 | | Release date: | 2017-01-25 | | Last modified: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Broad-Spectrum Kinase Profiling in Live Cells with Lysine-Targeted Sulfonyl Fluoride Probes.
J. Am. Chem. Soc., 139, 2017
|
|
8QWX
 
 | | Ligninolytic manganese peroxidase Ape-MnP1 from Agaricales mushrooms | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Santillana, E, Romero, A. | | Deposit date: | 2023-10-20 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-function characterization of two enzymes from novel subfamilies of manganese peroxidases secreted by the lignocellulose-degrading Agaricales fungi Agrocybe pediades and Cyathus striatus.
Biotechnol Biofuels Bioprod, 17, 2024
|
|
6PHL
 
 | |
7ZJK
 
 | | CspZ (BbCRASP-2) from Borrelia burgdorferi strain B408 | | Descriptor: | CspZ | | Authors: | Brangulis, K, Marcinkiewicz, A, Hart, T.M, Dupuis, A.P, Zamba Campero, M, Nowak, T.A, Stout, J.L, Akopjana, I, Kazaks, A, Bogans, J, Ciota, A.T, Kraiczy, P, Kolokotronis, S.O, Lin, Y.-P. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural evolution of an immune evasion determinant shapes pathogen host tropism.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7PPM
 
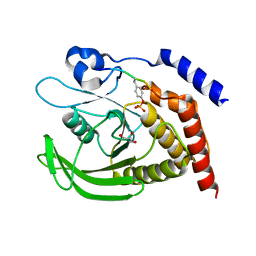 | | SHP2 catalytic domain in complex with IRS1 (889-901) phosphopeptide (pSer-892, pTyr-896) | | Descriptor: | GLYCEROL, Insulin receptor substrate 1, Tyrosine-protein phosphatase non-receptor type 11,Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Sok, P, Zeke, A, Remenyi, A. | | Deposit date: | 2021-09-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural insights into the pSer/pThr dependent regulation of the SHP2 tyrosine phosphatase in insulin and CD28 signaling.
Nat Commun, 13, 2022
|
|
6YC2
 
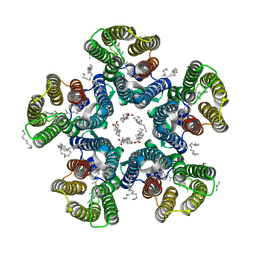 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric form at room temperature, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ALANINE, EICOSANE, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
7PPN
 
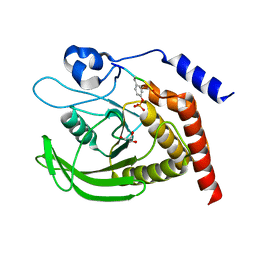 | | SHP2 catalytic domain in complex with CD28 (183-198) phosphopeptide (pTyr-191, p-Thr-195) | | Descriptor: | GLYCEROL, T-cell-specific surface glycoprotein CD28, Tyrosine-protein phosphatase non-receptor type 11,Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Sok, P, Zeke, A, Remenyi, A. | | Deposit date: | 2021-09-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the pSer/pThr dependent regulation of the SHP2 tyrosine phosphatase in insulin and CD28 signaling.
Nat Commun, 13, 2022
|
|
6EWG
 
 | |
6PHQ
 
 | |
6J0Y
 
 | | Crystal Structure of Yeast Rtt107 and Slx4 | | Descriptor: | Peptide from Structure-specific endonuclease subunit SLX4, Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
7L64
 
 | |
8RF3
 
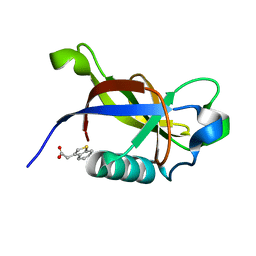 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7G3 refined against the anomalous diffraction data | | Descriptor: | 2-(1-benzothiophen-3-yl)ethanoic acid, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6J1J
 
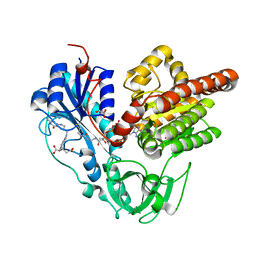 | | Crystal structure of HypX from Aquifex aeolicus, A392F-I419F variant in complex with Tetrahydrofolic acid | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, COENZYME A, GLYCEROL, ... | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of HypX responsible for CO biosynthesis in the maturation of NiFe-hydrogenase.
Commun Biol, 2, 2019
|
|
6EWJ
 
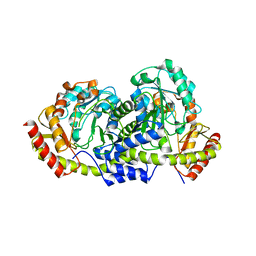 | | Putative sugar aminotransferase Spr1654 from Streptococcus pneumoniae, apo-form | | Descriptor: | Putative capsular polysaccharide biosynthesis protein | | Authors: | Achour, A, Sun, R, Sandalova, T, Han, X. | | Deposit date: | 2017-11-04 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional studies of Spr1654: an essential aminotransferase in teichoic acid biosynthesis inStreptococcus pneumoniae.
Open Biol, 8, 2018
|
|
5UA4
 
 | |
8QUS
 
 | |
7PHN
 
 | | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria | | Descriptor: | (2~{R},4~{R})-4-[[(2~{S},4~{S})-4-fluoranylpyrrolidin-2-yl]carbonylamino]-1-(2-methylpropanoyl)-~{N}-[[4-(2-phenylethynyl)phenyl]methyl]pyrrolidine-2-carboxamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Ryan, M.D, Pallin, T.D, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2021-08-17 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria
To Be Published
|
|
6PI1
 
 | |
7ZQZ
 
 | |
6EX9
 
 | |
6B7E
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with (R)-4-(5-(difluoromethyl)-1H-imidazol-1-yl)-3,3-dimethylisochroman-1-one | | Descriptor: | (4R)-4-[5-(difluoromethyl)-1H-imidazol-1-yl]-3,3-dimethyl-3,4-dihydro-1H-2-benzopyran-1-one, DIMETHYL SULFOXIDE, Phosphopantetheine adenylyltransferase, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
7KEP
 
 | | avibactam-CDD-1 2 minute complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (2S,5R)-7-oxo-6-(sulfooxy)-1,6-diazabicyclo[3.2.1]octane-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-10-11 | | Release date: | 2021-01-20 | | Last modified: | 2021-05-26 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Inhibition of the Clostridioides difficile Class D beta-Lactamase CDD-1 by Avibactam.
Acs Infect Dis., 7, 2021
|
|
5FU4
 
 | | The complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition | | Descriptor: | CBM74-RFGH5, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Basle, A, Luis, A.S, Venditto, I, Gilbert, H.J. | | Deposit date: | 2016-01-20 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
6PCD
 
 | |
7PJG
 
 | | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria | | Descriptor: | (2R,4R)-N-[[4-(4-cyclopropylbuta-1,3-diynyl)phenyl]methyl]-1-(2-methylpropanoyl)-4-[[(2S,4R)-4-oxidanylpyrrolidin-2-yl]carbonylamino]pyrrolidine-2-carboxamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Ryan, M.D, Pallin, T.D, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2021-08-24 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria
To Be Published
|
|
