8OKM
 
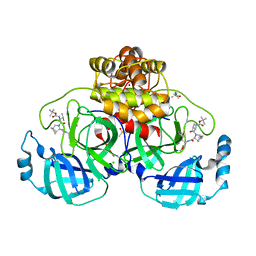 | | Crystal structure of F2F-2020197-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-03-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OPI
 
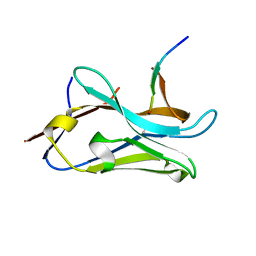 | | VHH Z70 mutant 1 in interaction with PHF6 Tau peptide | | Descriptor: | PHF6 Tau peptide, VHH Z70 mutant 1 | | Authors: | Dupre, E, Mortelecque, J, NGuyen, M, Hanoulle, X, Landrieu, I. | | Deposit date: | 2023-04-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A selection and optimization strategy for single-domain antibodies targeting the PHF6 linear peptide within the tau intrinsically disordered protein.
J.Biol.Chem., 300, 2024
|
|
5FC8
 
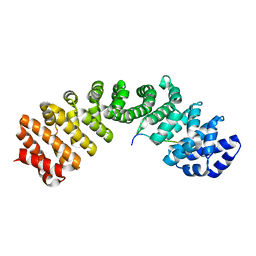 | | Mouse importin alpha: Dengue 3 NS5 C-terminal NLS peptide complex | | Descriptor: | Importin subunit alpha-1, Nonstructural protein 5 | | Authors: | Smith, K.M, Forwood, J.K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The C-terminal 18 Amino Acid Region of Dengue Virus NS5 Regulates its Subcellular Localization and Contains a Conserved Arginine Residue Essential for Infectious Virus Production.
PLoS Pathog., 12, 2016
|
|
8OKL
 
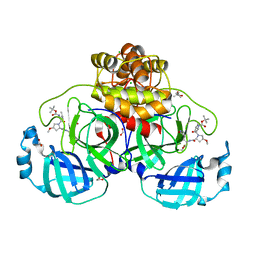 | | Crystal structure of F2F-2020185-01X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-03-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
5LQ1
 
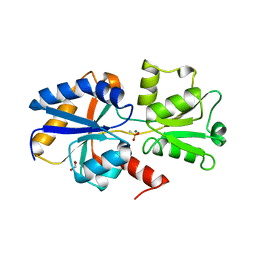 | | 1.41 A resolution structure of PtxB from Trichodesmium erythraeum IMS101 in complex with methylphosphonate | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, METHYLPHOSPHONIC ACID ESTER GROUP, ... | | Authors: | Bisson, C, Adams, N.B.P, Polyviou, D, Bibby, T.S, Hunter, C.N, Hitchcock, A. | | Deposit date: | 2016-08-15 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | The molecular basis of phosphite and hypophosphite recognition by ABC-transporters.
Nat Commun, 8, 2017
|
|
8P05
 
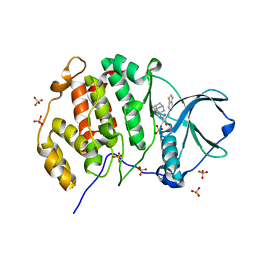 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with Leucettinib-92 | | Descriptor: | (4~{Z})-2-(1-adamantylamino)-4-(1,3-benzothiazol-6-ylmethylidene)-1~{H}-imidazol-5-one, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Kraemer, A, Meijer, L, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-09 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Leucettinibs, a Class of DYRK/CLK Kinase Inhibitors Inspired by the Marine Sponge Natural Product Leucettamine B.
J.Med.Chem., 66, 2023
|
|
8CV6
 
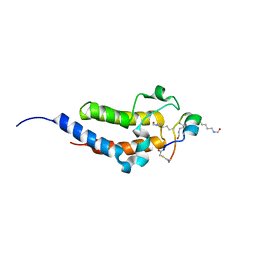 | | Peptide 4.2B in complex with BRD4.2 | | Descriptor: | ACETYL GROUP, AMINO GROUP, BRD4 protein, ... | | Authors: | Franck, C, Mackay, J.P. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and characterization of cyclic peptides selective for the C-terminal bromodomains of BET family proteins.
Structure, 31, 2023
|
|
8OKK
 
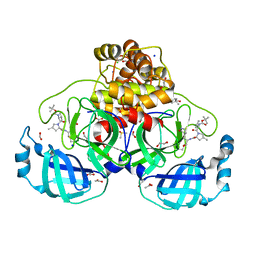 | | Crystal structure of F2F-2020184-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-03-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
5LR3
 
 | |
8P07
 
 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with 5-((3-(4H-1,2,4-triazol-4-yl)phenyl)amino)-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidine-3-carbonitrile | | Descriptor: | 1,2-ETHANEDIOL, 7-(cyclopropylamino)-5-[[3-(1,2,4-triazol-4-yl)phenyl]amino]pyrazolo[1,5-a]pyrimidine-3-carbonitrile, Casein kinase II subunit alpha, ... | | Authors: | Kraemer, A, Ong, H.W, Yang, X, Bown, J.W, Chang, E, Willson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | More than an Amide Bioisostere: Discovery of 1,2,4-Triazole-containing Pyrazolo[1,5- a ]pyrimidine Host CSNK2 Inhibitors for Combatting beta-Coronavirus Replication.
J.Med.Chem., 67, 2024
|
|
8D02
 
 | | Crystal Structure of Bacillus anthracis BxpB | | Descriptor: | BxpB, CALCIUM ION | | Authors: | Chattopadhyay, D, Turnbough, C.L. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure and induced stability of trimeric BxpB: implications for the assembly of BxpB-BclA complexes in the exosporium of Bacillus anthracis.
Mbio, 14, 2023
|
|
5LR5
 
 | |
8D1S
 
 | | Crystal structure of Plasmodium falciparum GRP78 in complex with Toyocamycin | | Descriptor: | 4-amino-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Chaperone DnaK, SULFATE ION | | Authors: | Mrozek, A, Chen, Y, Antoshchenko, T, Park, H.W. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A non-traditional crystal-based compound screening method targeting the ATP binding site of Plasmodium falciparum GRP78 for identification of novel nucleoside analogues.
Front Mol Biosci, 9, 2022
|
|
5LRQ
 
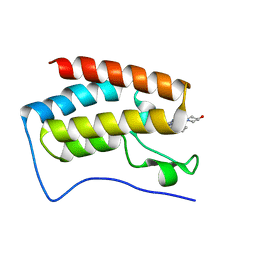 | | BRD4 in complex with ERK5 inhibitor XMD8-92 | | Descriptor: | 2-{[2-ethoxy-4-(4-hydroxypiperidin-1-yl)phenyl]amino}-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4 | | Authors: | Martin, M.P, Noble, M.E.M. | | Deposit date: | 2016-08-19 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of a novel orally bioavailable ERK5 inhibitor with selectivity over p38 alpha and BRD4.
Eur.J.Med.Chem., 178, 2019
|
|
5LRX
 
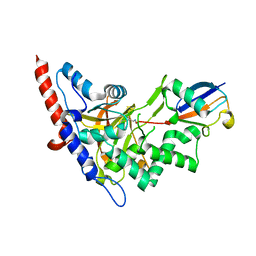 | | Structure of A20 OTU domain bound to ubiquitin | | Descriptor: | Polyubiquitin-B, Tumor necrosis factor alpha-induced protein 3 | | Authors: | Mevissen, T.E.T, Kulathu, Y, Mulder, M.P.C, Geurink, P.P, Maslen, S.L, Gersch, M, Elliott, P.R, Burke, J.E, van Tol, B.D.M, Akutsu, M, El Oualid, F, Kawasaki, M, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2016-08-22 | | Release date: | 2016-10-19 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Nature, 538, 2016
|
|
8D32
 
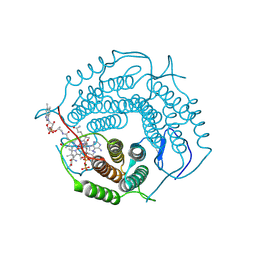 | | Mycobacterium tuberculosis pduO-type ATP:cobalamin adenosyltransferase bound to 5-deoxyadenosylrhodibalamin and PPPi | | Descriptor: | 5'-DEOXYADENOSINE, Corrinoid adenosyltransferase, MAGNESIUM ION, ... | | Authors: | Mascarenhas, R.N, Ruetz, M, Koutmos, M, Banerjee, R. | | Deposit date: | 2022-05-31 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A noble substitution leads to the cofactor mimicry by rhodibalamin
To Be Published
|
|
5FA5
 
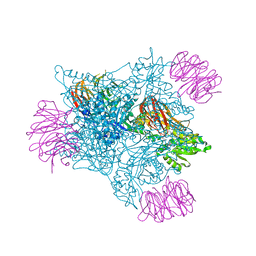 | |
5WBD
 
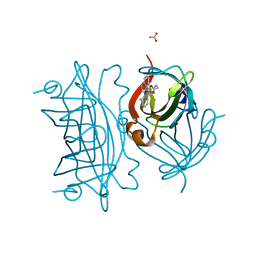 | | Peroxide Activation Regulated by Hydrogen Bonds within Artificial Cu Proteins - N49A | | Descriptor: | ACETATE ION, Streptavidin, [N-(2-{bis[2-(pyridin-2-yl-kappaN)ethyl]amino-kappaN}ethyl)-5-(2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentanamide](hydroxy)copper | | Authors: | Mann, S.I, Heinisch, T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peroxide Activation Regulated by Hydrogen Bonds within Artificial Cu Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
5LT3
 
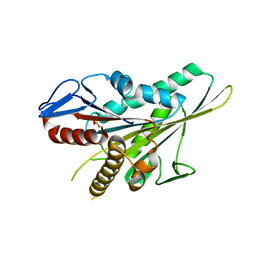 | |
8OKN
 
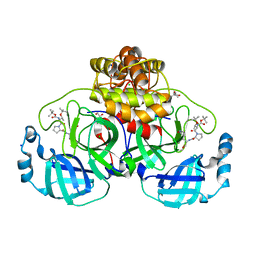 | | Crystal structure of F2F-2020198-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-03-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
5LU4
 
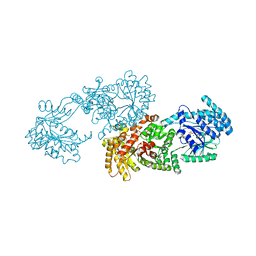 | | C4-type pyruvate phosphate dikinase: conformational intermediate of central domain in the swiveling mechanism | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PYRUVIC ACID, ... | | Authors: | Minges, A, Hoeppner, A, Groth, G. | | Deposit date: | 2016-09-08 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Trapped intermediate state of plant pyruvate phosphate dikinase indicates substeps in catalytic swiveling domain mechanism.
Protein Sci., 26, 2017
|
|
8P04
 
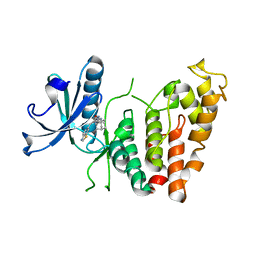 | | Crystal structure of human CLK1 in complex with Leucettinib-92 | | Descriptor: | (4~{Z})-2-(1-adamantylamino)-4-(1,3-benzothiazol-6-ylmethylidene)-1~{H}-imidazol-5-one, Dual specificity protein kinase CLK1 | | Authors: | Kraemer, A, Schroeder, M, Meijer, L, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-09 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Leucettinibs, a Class of DYRK/CLK Kinase Inhibitors Inspired by the Marine Sponge Natural Product Leucettamine B.
J.Med.Chem., 66, 2023
|
|
5LV0
 
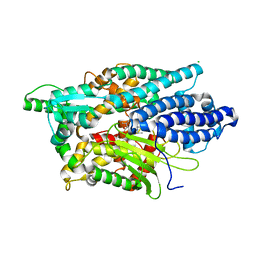 | | Structure of Human Neurolysin (E475Q) in complex with amyloid-beta 35-40 peptide product | | Descriptor: | CHLORIDE ION, GLY-VAL-VAL amyloid 35-40 fragment, Neurolysin, ... | | Authors: | Masuyer, G, Berntsson, R.P.-A, Teixeira, P.F, Kmiec, B, Glaser, E, Stenmark, P. | | Deposit date: | 2016-09-12 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional analysis of Neurolysin, a new component of the mitochondrial peptidolytic network
To Be Published
|
|
6N1E
 
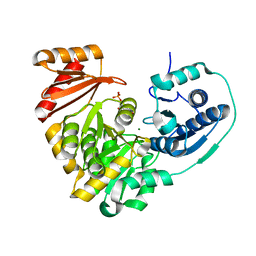 | | Crystal structure of X. citri phosphoglucomutase in complex with 1-methyl-glucose 6-phosphate | | Descriptor: | 1-deoxy-7-O-phosphono-alpha-D-gluco-hept-2-ulopyranose, MAGNESIUM ION, Phosphomannomutase/phosphoglucomutase | | Authors: | Beamer, L.J, Stiers, K.M. | | Deposit date: | 2018-11-08 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis, Derivatization, and Structural Analysis of Phosphorylated Mono-, Di-, and Trifluorinated d-Gluco-heptuloses by Glucokinase: Tunable Phosphoglucomutase Inhibition.
Acs Omega, 4, 2019
|
|
5W74
 
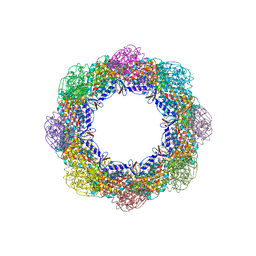 | | Crystal Structure of the Group II Chaperonin from Methanococcus Maripaludis D386ADeltaLid Mutant in the Open, ADP-Bound State | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin, MAGNESIUM ION | | Authors: | Dalton, K.M, Lopez, T, Liu, C, Ralston, C.Y, Pereira, J.H, Chartron, J.W, McAndrew, R.P, Douglas, N.R, Adams, P.D, Pande, V.S, Frydman, J. | | Deposit date: | 2017-06-19 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | The Conformational Cycle of the Group II Chaperonin Termini
To Be Published
|
|
