7FAG
 
 | | Room temperature structure of elastase with high-strength agarose hydrogel | | Descriptor: | CALCIUM ION, Chymotrypsin-like elastase family member 1, SULFATE ION, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Adachi, H, Murata, M, Mori, Y. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Room temperature structure of elastase with high-strength agarose hydrogel
To Be Published
|
|
3FVA
 
 | | NNQNTF segment from elk prion | | Descriptor: | Major prion protein | | Authors: | Apostol, M.I, Eisenberg, D. | | Deposit date: | 2009-01-15 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.458 Å) | | Cite: | Molecular mechanisms for protein-encoded inheritance.
Nat.Struct.Mol.Biol., 16, 2009
|
|
1ELV
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN COMPLEMENT C1S PROTEASE | | Descriptor: | 2-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C1S COMPONENT, ... | | Authors: | Gaboriaud, C, Rossi, V, Bally, I, Arlaud, G, Fontecilla-Camps, J.-C. | | Deposit date: | 2000-03-14 | | Release date: | 2001-03-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the catalytic domain of human complement c1s: a serine protease with a handle.
EMBO J., 19, 2000
|
|
3BWM
 
 | | Crystal Structure of Human Catechol O-Methyltransferase with bound SAM and DNC | | Descriptor: | 3,5-DINITROCATECHOL, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Rutherford, K, Le Trong, I, Stenkamp, R.E, Parson, W.W. | | Deposit date: | 2008-01-09 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of human 108V and 108M catechol O-methyltransferase.
J.Mol.Biol., 380, 2008
|
|
3CLN
 
 | |
3FR1
 
 | |
8TLY
 
 | |
8TUK
 
 | | Alvinella ASCC1 KH and Phosphodiesterase/Ligase Domain | | Descriptor: | 1,2-ETHANEDIOL, Activating signal cointegrator 1 complex subunit 1, IMIDAZOLE | | Authors: | Tsutakawa, S.E, Tainer, J.A, Arvai, A.S, Chinnam, N.B. | | Deposit date: | 2023-08-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | ASCC1 structures and bioinformatics reveal a novel helix-clasp-helix RNA-binding motif linked to a two-histidine phosphodiesterase.
J.Biol.Chem., 300, 2024
|
|
1SI5
 
 | | Protease-like domain from 2-chain hepatocyte growth factor | | Descriptor: | hepatocyte growth factor | | Authors: | Kirchhofer, D, Yao, X, Peek, M, Eigenbrot, C, Lipari, M.T, Billeci, K.L, Maun, H.R, Moran, P, Santell, L, Lazarus, R.A. | | Deposit date: | 2004-02-27 | | Release date: | 2004-12-28 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural and functional basis of the serine protease-like hepatocyte growth factor beta-chain in Met binding and signaling
J.Biol.Chem., 279, 2004
|
|
5SGA
 
 | |
8A7P
 
 | |
4K1T
 
 | | Gly-Ser-SplB protease from Staphylococcus aureus at 1.60 A resolution | | Descriptor: | CHLORIDE ION, SULFATE ION, Serine protease SplB, ... | | Authors: | Zdzalik, M, Pustelny, K, Stec-Niemczyk, J, Cichon, P, Czarna, A, Popowicz, G, Drag, M, Wladyka, B, Potempa, J, Dubin, A, Dubin, G. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Staphylococcal SplB Serine Protease Utilizes a Novel Molecular Mechanism of Activation.
J.Biol.Chem., 289, 2014
|
|
4K1S
 
 | | Gly-Ser-SplB protease from Staphylococcus aureus at 1.96 A resolution | | Descriptor: | Serine protease SplB | | Authors: | Zdzalik, M, Pustelny, K, Stec-Niemczyk, J, Cichon, P, Czarna, A, Popowicz, G, Drag, M, Wladyka, B, Potempa, J, Dubin, A, Dubin, G. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Staphylococcal SplB Serine Protease Utilizes a Novel Molecular Mechanism of Activation.
J.Biol.Chem., 289, 2014
|
|
1HLT
 
 | |
6EST
 
 | |
6TNA
 
 | | CRYSTAL STRUCTURE OF YEAST PHENYLALANINE T-RNA. I.CRYSTALLOGRAPHIC REFINEMENT | | Descriptor: | MAGNESIUM ION, TRNAPHE | | Authors: | Sussman, J.L, Holbrook, S.R, Warrant, R.W, Church, G.M, Kim, S.-H. | | Deposit date: | 1978-11-16 | | Release date: | 1979-01-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement.
J.Mol.Biol., 123, 1978
|
|
6TH7
 
 | |
8FNJ
 
 | | Structure of E138K/G140A HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNN
 
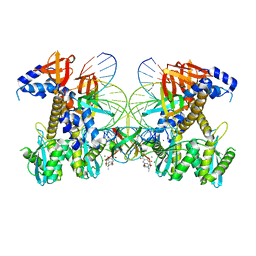 | | Structure of E138K/G140A/Q148K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FND
 
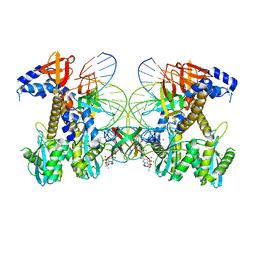 | | Structure of E138K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNO
 
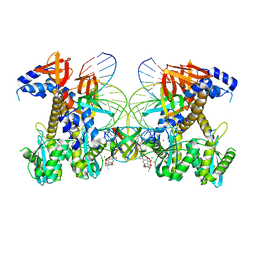 | | Structure of E138K/G140A/Q148R HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-28 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FN7
 
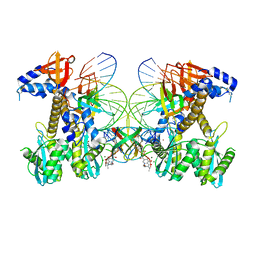 | | Structure of WT HIV-1 intasome bound to Dolutegravir | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNH
 
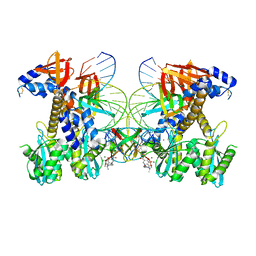 | | Structure of Q148K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNL
 
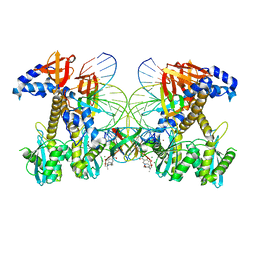 | | Structure of E138K/Q148K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNP
 
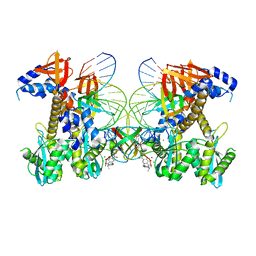 | | Structure of E138K/G140S/Q148H HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
