7KO4
 
 | |
8C8H
 
 | |
7KO5
 
 | |
7KOR
 
 | |
4WRC
 
 | | Crystal Structure of Surfactant Protein-A DEDN Mutant (E171D/P175E/R197N/K203D) | | Descriptor: | CALCIUM ION, CHLORIDE ION, HEXANE-1,6-DIOL, ... | | Authors: | Rynkiewicz, M.J, Wu, H, Cafarella, T.R, Nikolaidis, N.M, Head, J.F, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2014-10-23 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Differential ligand binding specificities of the pulmonary collectins are determined by the conformational freedom of a surface loop
to be published
|
|
6L7A
 
 | | CsgFG complex in Curli biogenesis system | | Descriptor: | CsgF, Curli production assembly/transport protein CsgG | | Authors: | Yan, Z.F, Yin, M, Chen, J.N, Li, X.M. | | Deposit date: | 2019-11-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Assembly and substrate recognition of curli biogenesis system.
Nat Commun, 11, 2020
|
|
1IPP
 
 | | HOMING ENDONUCLEASE/DNA COMPLEX | | Descriptor: | CADMIUM ION, DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*AP*GP*AP*GP*AP*GP*TP*CP*A)-3'), INTRON-ENCODED ENDONUCLEASE I-PPOI, ... | | Authors: | Flick, K.E, Jurica, M.S, Monnat Jr, R.J, Stoddard, B.L. | | Deposit date: | 1998-03-19 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI.
Nature, 394, 1998
|
|
7KO7
 
 | |
2JJM
 
 | |
4X0C
 
 | |
7KON
 
 | |
6KQE
 
 | | Thermus thermophilus initial transcription complex comprising sigma A and 5'-OH RNA of 4 nt | | Descriptor: | DNA (5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G*)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, Y, Li, L, Ebright, R.H. | | Deposit date: | 2019-08-17 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1IUT
 
 | | P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE AT PH 7.4 | | Descriptor: | 4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Gatti, D.L, Entsch, B, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1995-11-22 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis.
Biochemistry, 35, 1996
|
|
8C58
 
 | | CpG specific M.MpeI methyltransferase crystallized in the presence of 5-hydroxycytosine and 5-methylcytosine containing dsDNA | | Descriptor: | CARBONATE ION, Cytosine-specific methyltransferase, DNA (5'-D(*CP*CP*AP*CP*AP*TP*GP*(5OC)P*GP*CP*TP*GP*AP*A)-3'), ... | | Authors: | Wojciechowski, M, Czapinska, H, Krwawicz, J, Rafalski, D, Bochtler, M. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cytosine analogues as DNA methyltransferase substrates.
Nucleic Acids Res., 52, 2024
|
|
4X1W
 
 | | Crystal structure of unbound RHDVb P domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, VP1 | | Authors: | Leuthold, M.M, Hansman, G.S. | | Deposit date: | 2014-11-25 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of a rabbit hemorrhagic disease virus binding to histo-blood group antigens.
J.Virol., 89, 2015
|
|
1IVH
 
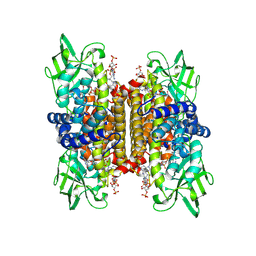 | | STRUCTURE OF HUMAN ISOVALERYL-COA DEHYDROGENASE AT 2.6 ANGSTROMS RESOLUTION: STRUCTURAL BASIS FOR SUBSTRATE SPECIFICITY | | Descriptor: | COENZYME A PERSULFIDE, FLAVIN-ADENINE DINUCLEOTIDE, ISOVALERYL-COA DEHYDROGENASE | | Authors: | Tiffany, K.A, Roberts, D.L, Wang, M, Paschke, R, Mohsen, A.-W.A, Vockley, J, Kim, J.J.P. | | Deposit date: | 1997-05-15 | | Release date: | 1998-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human isovaleryl-CoA dehydrogenase at 2.6 A resolution: structural basis for substrate specificity,.
Biochemistry, 36, 1997
|
|
8C59
 
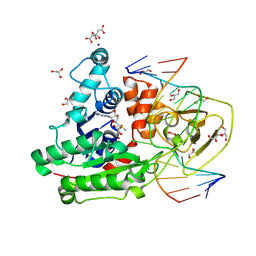 | | CpG specific M.MpeI methyltransferase crystallized in the presence of 5-bromocytosine (converted to 5mC) and 5-methylcytosine containing dsDNA | | Descriptor: | CARBONATE ION, CITRIC ACID, Cytosine-specific methyltransferase, ... | | Authors: | Wojciechowski, M, Czapinska, H, Krwawicz, J, Rafalski, D, Bochtler, M. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cytosine analogues as DNA methyltransferase substrates.
Nucleic Acids Res., 52, 2024
|
|
2JDX
 
 | |
1IT5
 
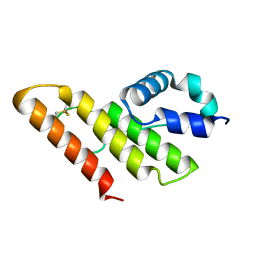 | | Solution structure of apo-type PLA2 from Streptomyces violaceruber A-2688. | | Descriptor: | Phospholipase A2 | | Authors: | Sugiyama, M, Ohtani, K, Izuhara, M, Koike, T. | | Deposit date: | 2002-01-09 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel prokaryotic phospholipase A2. Characterization, gene cloning, and solution structure.
J.Biol.Chem., 277, 2002
|
|
8C57
 
 | | CpG specific M.MpeI methyltransferase crystallized in the presence of 5,6-dihydro-5-azacytosine (converted to 5m-dhaC) and 5-methylcytosine containing dsDNA | | Descriptor: | CARBONATE ION, Cytosine-specific methyltransferase, DNA (5'-D(*CP*CP*AP*CP*AP*TP*GP*(5MA)P*GP*CP*TP*GP*AP*A)-3'), ... | | Authors: | Wojciechowski, M, Czapinska, H, Krwawicz, J, Rafalski, D, Bochtler, M. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cytosine analogues as DNA methyltransferase substrates.
Nucleic Acids Res., 52, 2024
|
|
1IWK
 
 | | Putidaredoxin-Binding Stablilizes an Active Conformer of Cytochrome P450cam in its Reduced State; Crystal Structure of Mutant(112K) Cytochrome P450cam | | Descriptor: | CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Shimada, H, Tarumi, A, Hishiki, T, Kimata-Ariga, Y, Egawa, T, Park, S.-Y, Adachi, S, Shiro, Y, Ishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-05-15 | | Release date: | 2002-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Infrared spectroscopic and mutational studies on putidaredoxin-induced conformational changes in ferrous CO-P450cam
Biochemistry, 42, 2003
|
|
6DTI
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with an unmodifed anticodon stem loop (ASL) of Escherichia coli transfer RNA Arginine 2 (TRNAARG2) bound to an mRNA with an CGU-codon in the A-site and paromomycin | | Descriptor: | 16s rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Cantara, W.A, DeMirci, H, Agris, P.F. | | Deposit date: | 2018-06-16 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | A Structural Basis for Restricted Codon Recognition Mediated by 2-thiocytidine in tRNA Containing a Wobble Position Inosine.
J.Mol.Biol., 432, 2020
|
|
1J1C
 
 | | Binary complex structure of human tau protein kinase I with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glycogen synthase kinase-3 beta, MAGNESIUM ION | | Authors: | Aoki, M, Yokota, T, Sugiura, I, Sasaki, C, Hasegawa, T, Okumura, C, Kohno, T, Sugio, S, Matsuzaki, T. | | Deposit date: | 2002-12-03 | | Release date: | 2003-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into nucleotide recognition in tau-protein kinase I/glycogen synthase kinase 3 beta.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4X83
 
 | | Crystal structure of Dscam1 isoform 7.44, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4WKG
 
 | |
