8F0P
 
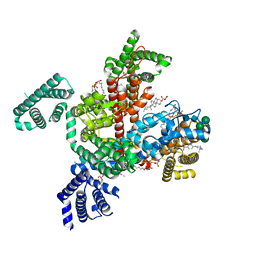 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the hybrid inhibitor GNE-1305 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
8ONA
 
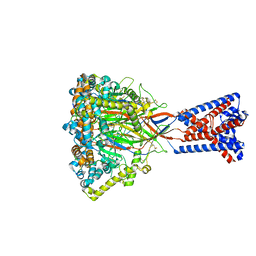 | | FMRFa-bound Malacoceros FaNaC1 in lipid nanodiscs in presence of diminazene | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FMRFamide, neuropeptide, ... | | Authors: | Kalienkova, V, Dandamudi, M, Paulino, C, Lynagh, T. | | Deposit date: | 2023-04-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for excitatory neuropeptide signaling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8F0R
 
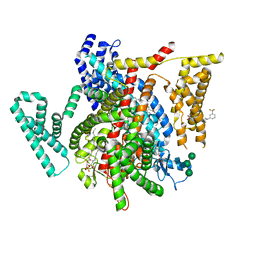 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the arylsulfonamide inhibitor GNE-3565 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloro-4-({(1S,2S,4S)-2-(dimethylamino)-4-[3-(trifluoromethyl)phenyl]cyclohexyl}amino)-2-fluoro-N-(pyrimidin-4-yl)benzene-1-sulfonamide, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
6XYR
 
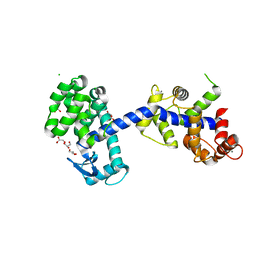 | | Structure of the T4Lnano fusion protein | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Benoit, R.M, Bierig, T, Collu, C, Engilberge, S, Olieric, V. | | Deposit date: | 2020-01-31 | | Release date: | 2020-12-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Chimeric single alpha-helical domains as rigid fusion protein connections for protein nanotechnology and structural biology.
Structure, 30, 2022
|
|
8PAT
 
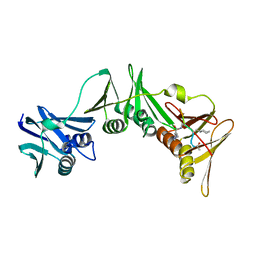 | | Structure of the E.coli DNA polymerase sliding clamp with a covalently bound peptide 3. | | Descriptor: | ACE-GLN-ALC-GLX-LEU-PHE, Beta sliding clamp | | Authors: | Compain, G, Monsarrat, C, Blagojevic, J, Brillet, K, Dumas, P, Hammann, P, Kuhn, L, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Burnouf, D, Guichard, G. | | Deposit date: | 2023-06-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Peptide-Based Covalent Inhibitors Bearing Mild Electrophiles to Target a Conserved His Residue of the Bacterial Sliding Clamp.
Jacs Au, 4, 2024
|
|
5LKC
 
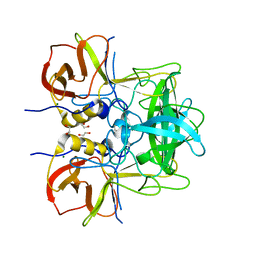 | | Protruding domain of GII.17 norovirus Kawasaki308 in complex with HBGA type A (triglycan) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Major capsid protein, ... | | Authors: | Singh, B.K, Morozov, V, Hansman, G.S. | | Deposit date: | 2016-07-22 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Human norovirus inhibition by a human milk oligosaccharide.
Virology, 508, 2017
|
|
5LKG
 
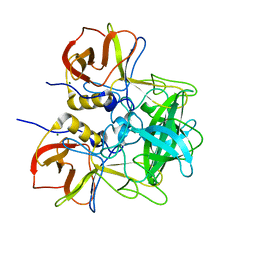 | |
6XQM
 
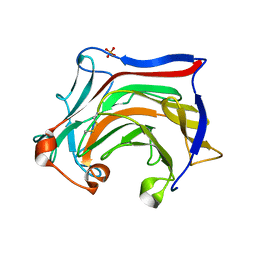 | | Crystal structure of SCLam E144S mutant, a non-specific endo-beta-1,3(4)-glucanase from family GH16, co-crystallized with laminarihexaose, presenting a laminaribiose and a glucose at active site | | Descriptor: | CALCIUM ION, GH16 family protein, PHOSPHATE ION, ... | | Authors: | Liberato, M.V, Squina, F. | | Deposit date: | 2020-07-09 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insights into the dual cleavage activity of the GH16 laminarinase enzyme class on beta-1,3 and beta-1,4 glycosidic bonds.
J.Biol.Chem., 296, 2021
|
|
6XQH
 
 | | Crystal structure of SCLam E144S mutant, a non-specific endo-beta-1,3(4)-glucanase from family GH16, co-crystallized with cellotriose, presenting a 1,3-beta-D-cellobiosyl-glucose and a cellobiose at active site | | Descriptor: | CALCIUM ION, GH16 family protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Liberato, M.V, Squina, F. | | Deposit date: | 2020-07-09 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Insights into the dual cleavage activity of the GH16 laminarinase enzyme class on beta-1,3 and beta-1,4 glycosidic bonds.
J.Biol.Chem., 296, 2021
|
|
6XQG
 
 | | Crystal structure of SCLam E144S mutant, a non-specific endo-beta-1,3(4)-glucanase from family GH16, co-crystallized with 1,3-beta-D-cellobiosyl-cellobiose, presenting a 1,3-beta-D-cellobiosyl-glucose at active site | | Descriptor: | CALCIUM ION, GH16 family protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Liberato, M.V, Squina, F. | | Deposit date: | 2020-07-09 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insights into the dual cleavage activity of the GH16 laminarinase enzyme class on beta-1,3 and beta-1,4 glycosidic bonds.
J.Biol.Chem., 296, 2021
|
|
6P50
 
 | | Crystal Structure of a Complex of human IL-7Ralpha with an anti-IL-7Ralpha Fab 4A10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-7 receptor subunit alpha, anti-IL-7R 4A10 Fab heavy chain, ... | | Authors: | Walsh, S.T.R, Kashi, L, Kohnhorst, C.L. | | Deposit date: | 2019-05-29 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | New anti-IL-7R alpha monoclonal antibodies show efficacy against T cell acute lymphoblastic leukemia in pre-clinical models.
Leukemia, 34, 2020
|
|
6XQL
 
 | | Crystal structure of SCLam E144S mutant, a non-specific endo-beta-1,3(4)-glucanase from family GH16, co-crystallized with cellohexaose, presenting a 1,3-beta-D-cellobiosyl-glucose at active site | | Descriptor: | CALCIUM ION, GH16 family protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Liberato, M.V, Squina, F. | | Deposit date: | 2020-07-09 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Insights into the dual cleavage activity of the GH16 laminarinase enzyme class on beta-1,3 and beta-1,4 glycosidic bonds.
J.Biol.Chem., 296, 2021
|
|
8P36
 
 | | Neisseria meningitidis Type IV pilus SB-DATDH variant | | Descriptor: | 2,4-bisacetamido-2,4,6-trideoxy-beta-D-glucopyranose, Neisseria meningitidis PilE, SB-DATDH variant, ... | | Authors: | Fernandez-Martinez, D, Dumenil, G. | | Deposit date: | 2023-05-17 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Cryo-EM structures of type IV pili complexed with nanobodies reveal immune escape mechanisms.
Nat Commun, 15, 2024
|
|
3NJP
 
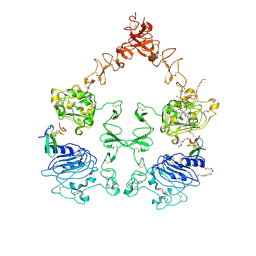 | | The Extracellular and Transmembrane Domain Interfaces in Epidermal Growth Factor Receptor Signaling | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor, Epidermal growth factor receptor, ... | | Authors: | Lu, C, Mi, L.-Z, Grey, M.J, Zhu, J, Graef, E, Yokoyama, S, Springer, T.A. | | Deposit date: | 2010-06-17 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.304 Å) | | Cite: | Structural evidence for loose linkage between ligand binding and kinase activation in the epidermal growth factor receptor.
Mol.Cell.Biol., 30, 2010
|
|
8PDY
 
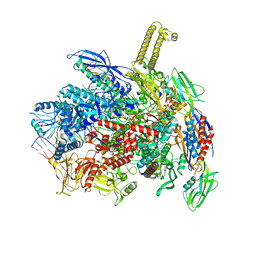 | | E. coli RNA polymerase paused at ops site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuber, P.K, Said, N, Hilal, T, Loll, B, Wahl, M.C, Knauer, S.H. | | Deposit date: | 2023-06-13 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Concerted transformation of a hyper-paused transcription complex and its reinforcing protein.
Nat Commun, 15, 2024
|
|
8P7C
 
 | |
6XPO
 
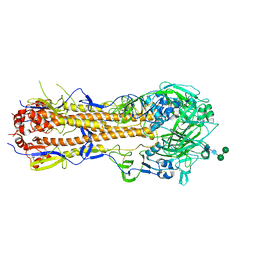 | | Influenza hemagglutinin A/Bangkok/01/1979(H3N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | McCarthy, K.R, Harrison, S.C. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Prevalent Focused Human Antibody Response to the Influenza Virus Hemagglutinin Head Interface.
Mbio, 12, 2021
|
|
8PE2
 
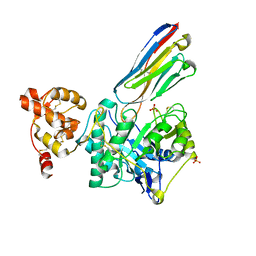 | | Crystal structure of Gel4 in complex with Nanobody 3 | | Descriptor: | 1,3-beta-glucanosyltransferase, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody 3, ... | | Authors: | Macias-Leon, J, Redrado-Hernandez, S, Castro-Lopez, J, Sanz, A.B, Arias, M, Farkas, V, Vincke, C, Muyldermans, S, Pardo, J, Arroyo, J, Galvez, E, Hurtado-Guerrero, R. | | Deposit date: | 2023-06-13 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Broad Protection against Invasive Fungal Disease from a Nanobody Targeting the Active Site of Fungal beta-1,3-Glucanosyltransferases.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8P4N
 
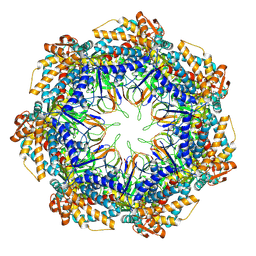 | | CryoEM structure of a GroEL7-GroES7 cage with encapsulated disordered substrate MetK in the presence of ADP-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Beck, F, Bracher, A, Caravajal, A.I, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography
Nature, 2024
|
|
8PE1
 
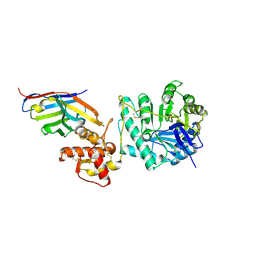 | | Crystal structure of Gel4 in complex with Nanobody 4 | | Descriptor: | 1,3-beta-glucanosyltransferase, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody 4, ... | | Authors: | Macias-Leon, J, Redrado-Hernandez, S, Castro-Lopez, J, Sanz, A.B, Arias, M, Farkas, V, Vincke, C, Muyldermans, S, Pardo, J, Arroyo, J, Galvez, E, Hurtado-Guerrero, R. | | Deposit date: | 2023-06-13 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Broad Protection against Invasive Fungal Disease from a Nanobody Targeting the Active Site of Fungal beta-1,3-Glucanosyltransferases.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8OMJ
 
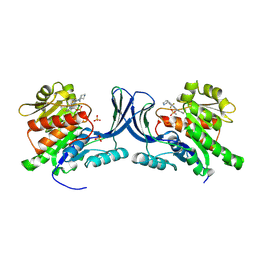 | | hKHK-C in complex with compound 28 | | Descriptor: | Ketohexokinase, SULFATE ION, [3-[[6-[(3~{a}~{R},6~{a}~{S})-2,3,3~{a},4,6,6~{a}-hexahydro-1~{H}-pyrrolo[3,4-c]pyrrol-5-yl]-3-cyano-4-(trifluoromethyl)pyridin-2-yl]amino]-4-methylsulfanyl-phenyl]methoxy-methyl-phosphinic acid | | Authors: | Pautsch, A, Ebenhoch, R. | | Deposit date: | 2023-03-31 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Discovery of BI-9787, a Potent Zwitterionic KHK-Inhibitor with Oral Bioavailability
To Be Published
|
|
8OPS
 
 | |
8OPT
 
 | |
6U7F
 
 | | HCoV-229E RBD Class IV in complex with human APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tomlinson, A.C.A, Li, Z, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
5I81
 
 | | aSMase with zinc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Zhou, Y.F, Wei, R.R. | | Deposit date: | 2016-02-18 | | Release date: | 2016-09-07 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Human acid sphingomyelinase structures provide insight to molecular basis of Niemann-Pick disease.
Nat Commun, 7, 2016
|
|
