4JQU
 
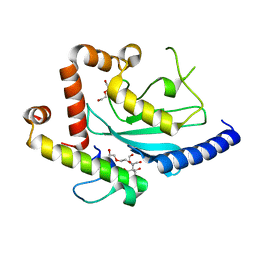 | | Crystal structure of Ubc7p in complex with the U7BR of Cue1p | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Coupling of ubiquitin conjugation to ER degradation protein 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liang, Y.-H, Metzger, M.B, Weissman, A.M, Ji, X. | | Deposit date: | 2013-03-20 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A Structurally Unique E2-Binding Domain Activates Ubiquitination by the ERAD E2, Ubc7p, through Multiple Mechanisms.
Mol.Cell, 50, 2013
|
|
1UIY
 
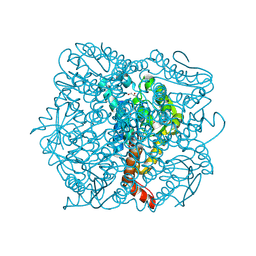 | | Crystal Structure of Enoyl-CoA Hydratase from Thermus Thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Enoyl-CoA Hydratase, GLYCEROL | | Authors: | Bagautdinov, B, Kuramitsu, S, Yokoyama, S, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-24 | | Release date: | 2003-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of enoyl-CoA hydratase from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
8B6C
 
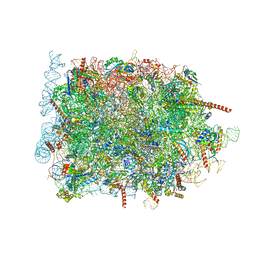 | | Cryo-EM structure of ribosome-Sec61 in complex with cyclotriazadisulfonamide derivative CK147 | | Descriptor: | 28S rRNA, 4-[[9-(cyclohexylmethyl)-3-methylidene-5-(4-methylphenyl)sulfonyl-1,5,9-triazacyclododec-1-yl]sulfonyl]-~{N},~{N}-dimethyl-aniline, 5.8S rRNA, ... | | Authors: | Pauwels, E, Shewakramani, N.R, De Wijngaert, B, Vermeire, K, Das, K. | | Deposit date: | 2022-09-26 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural insights into TRAP association with ribosome-Sec61 complex and translocon inhibition by a CADA derivative.
Sci Adv, 9, 2023
|
|
2N3B
 
 | | Structure of oxidized horse heart cytochrome c encapsulated in reverse micelles | | Descriptor: | Cytochrome c, HEME C | | Authors: | O'Brien, E.S, Nucci, N.V, Fuglestad, B, Tommos, C, Wand, A. | | Deposit date: | 2015-05-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Defining the Apoptotic Trigger: THE INTERACTION OF CYTOCHROME c AND CARDIOLIPIN.
J.Biol.Chem., 290, 2015
|
|
8BRP
 
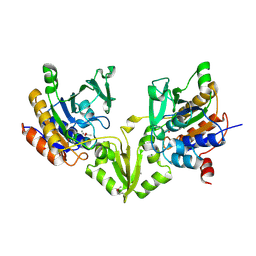 | | Peptide Arginase OspR from the cyanobacterium Kamptonema sp. | | Descriptor: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Mordhorst, S, Badmann, T, Boesch, N.M, Morinaka, B.I, Rach, H, Piel, J, Groll, M, Vagstadt, A.L. | | Deposit date: | 2022-11-23 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Biochemical Insights into Post-Translational Arginine-to-Ornithine Peptide Modifications by an Atypical Arginase.
Acs Chem.Biol., 18, 2023
|
|
7LL8
 
 | | D-Protein RFX-V1 Bound to the VEGFR1 Domain 2 Site on VEGF-A | | Descriptor: | Isoform L-VEGF189 of Vascular endothelial growth factor A, RFX-V1 | | Authors: | Marinec, P.S, Landgraf, K.E, Uppalapati, M, Chen, G, Xie, D, Jiang, Q, Zhao, Y, Petriello, A, Deshayes, K, Kent, S.B.H, Ault-Riche, D, Sidhu, S.S. | | Deposit date: | 2021-02-03 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A Non-immunogenic Bivalent d-Protein Potently Inhibits Retinal Vascularization and Tumor Growth.
Acs Chem.Biol., 16, 2021
|
|
8B5L
 
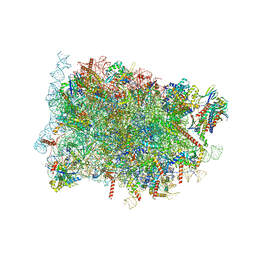 | | Cryo-EM structure of ribosome-Sec61-TRAP (TRanslocon Associated Protein) translocon complex | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Pauwels, E, Shewakramani, N.R, De Wijngaert, B, Vermeire, K, Das, K. | | Deposit date: | 2022-09-23 | | Release date: | 2023-03-01 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural insights into TRAP association with ribosome-Sec61 complex and translocon inhibition by a CADA derivative.
Sci Adv, 9, 2023
|
|
7LL9
 
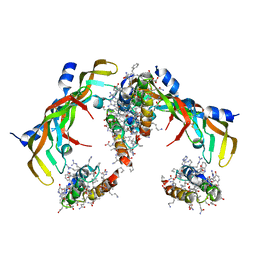 | | D-Protein RFX-V2 Bound to the VEGFR1 Domain 3 Site on VEGF-A | | Descriptor: | Isoform L-VEGF189 of Vascular endothelial growth factor A, RFX-V2 | | Authors: | Marinec, P.S, Landgraf, K.E, Uppalapati, M, Chen, G, Xie, D, Jiang, Q, Zhao, Y, Petriello, A, Deshayes, K, Kent, S.B.H, Ault-Riche, D, Sidhu, S.S. | | Deposit date: | 2021-02-03 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Non-immunogenic Bivalent d-Protein Potently Inhibits Retinal Vascularization and Tumor Growth.
Acs Chem.Biol., 16, 2021
|
|
3F8T
 
 | |
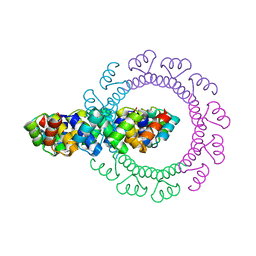
4YXY
 
 | |
2NCD
 
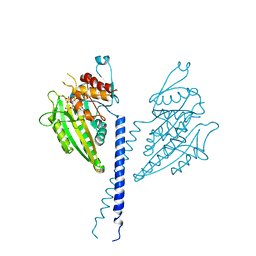 | | NCD (NON-CLARET DISJUNCTIONAL) DIMER FROM D. MELANOGASTER | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PROTEIN (Kinesin motor NCD), SULFATE ION | | Authors: | Sablin, E.P, Case, R.B, Dai, S.C, Hart, C.L, Ruby, A, Vale, R.D, Fletterick, R.J. | | Deposit date: | 1999-06-04 | | Release date: | 1999-06-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Direction determination in the minus-end-directed kinesin motor ncd.
Nature, 395, 1998
|
|
8BCH
 
 | | Human Brr2 Helicase Region in complex with Sulfaguanidine | | Descriptor: | 1-(4-aminophenyl)sulfonylguanidine, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Vester, K, Loll, B, Wahl, M.C. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Conformation-dependent ligand hot spots in the spliceosomal RNA helicase BRR2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6XNV
 
 | | CRYSTAL STRUCTURE OF LISTERIA MONOCYTOGENES CBPB PROTEIN (LMO1009) IN COMPLEX WITH C-DI-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CBS domain-containing protein | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2020-07-04 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | (p)ppGpp and c-di-AMP Homeostasis Is Controlled by CbpB in Listeria monocytogenes.
Mbio, 11, 2020
|
|
2MVX
 
 | | Atomic-resolution 3D structure of amyloid-beta fibrils: the Osaka mutation | | Descriptor: | Amyloid beta A4 protein | | Authors: | Schuetz, A.K, Vagt, T, Huber, M, Ovchinnikova, O.Y, Cadalbert, R, Wall, J, Guentert, P, Bockmann, A, Glockshuber, R, Meier, B.H. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-Resolution Three-Dimensional Structure of Amyloid beta Fibrils Bearing the Osaka Mutation.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1UJZ
 
 | | Crystal structure of the E7_C/Im7_C complex; a computationally designed interface between the colicin E7 DNase and the Im7 Immunity protein | | Descriptor: | Designed Colicin E7 DNase, Designed Colicin E7 immunity protein | | Authors: | Kortemme, T, Joachimiak, L.A, Bullock, A.N, Schuler, A.D, Stoddard, B.L, Baker, D. | | Deposit date: | 2003-08-13 | | Release date: | 2004-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Computational redesign of protein-protein interaction specificity
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
3FB7
 
 | | Open KcsA potassium channel in the presence of Rb+ ion | | Descriptor: | RUBIDIUM ION, Voltage-gated potassium channel, antibody fab fragment heavy chain, ... | | Authors: | Cuello, L.G, Jogini, V, Cortes, D.M, Pan, A.C, Gagnon, D.H, Cordero-Morales, J.F, Chakrapani, S, Roux, B, Perozo, E. | | Deposit date: | 2008-11-18 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Open KcsA potassium channel in the presence of Rb+ ion
TO BE PUBLISHED
|
|
2NND
 
 | | The Structural Identification of the Interaction Site and Functional State of RBP for its Membrane Receptor | | Descriptor: | 2-ISOBUTYL-3-METHOXYPYRAZINE, CADMIUM ION, Major urinary protein 2 | | Authors: | Redondo, C, Bingham, R.J, Vouropoulou, M, Homans, S.W, Findlay, J.B. | | Deposit date: | 2006-10-24 | | Release date: | 2007-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of the retinol-binding protein (RBP) interaction site and functional state of RBPs for the membrane receptor.
Faseb J., 22, 2008
|
|
4ZE2
 
 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) Y140H mutant complexed with itraconazole | | Descriptor: | 2-[(2R)-butan-2-yl]-4-{4-[4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazin-1-yl]phenyl}-2,4-dihydro-3H-1,2,4-triazol-3-one, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R, Monk, B.C, Tyndall, J.D.A. | | Deposit date: | 2015-04-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Triazole resistance mediated by mutations of a conserved active site tyrosine in fungal lanosterol 14 alpha-demethylase.
Sci Rep, 6, 2016
|
|
4L9B
 
 | | Structure of native RBP from lactococcal phage 1358 (CsI derivative) | | Descriptor: | CESIUM ION, Receptor Binding Protein | | Authors: | Farenc, C, Spinelli, S, Bebeacua, C, Tremblay, D, Orlov, I, Blangy, S, Klaholz, B.P, Moineau, S, Cambillau, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Virulent Siphophage CyoEM Structure and Host Recognition and Infection Mechanism
To be Published
|
|
8UK5
 
 | | Crystal structure of the bromodomain of human ATAD2B in complex with histone H4S1(ph)K5ac | | Descriptor: | ATPase family AAA domain-containing protein 2B, Histone H4S1(ph)K5ac | | Authors: | Montgomery, C, Phillips, M, Nix, J.C, Glass, K.C. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Impact of Combinatorial Histone Modifications on Acetyllysine Recognition by the ATAD2 and ATAD2B Bromodomains.
J.Med.Chem., 67, 2024
|
|
8OIG
 
 | | Crystal Structure of Staphopain C from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | McEwen, A.G, Magoch, M, Napolitano, V, Dubin, G, Wladyka, B. | | Deposit date: | 2023-03-22 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Staphopain C from Staphylococcus aureus.
Molecules, 28, 2023
|
|
5NX0
 
 | | Structure of Spin-labelled T4 lysozyme mutant L115C-R119C-R1 at room temperature | | Descriptor: | Endolysin | | Authors: | Gohlke, U, Loll, B, Consentius, P, Mueller, R, Kaupp, M, Heinemann, U, Wahl, M.C, Risse, T. | | Deposit date: | 2017-05-09 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Combining EPR spectroscopy and X-ray crystallography to elucidate the structure and dynamics of conformationally constrained spin labels in T4 lysozyme single crystals.
Phys Chem Chem Phys, 19, 2017
|
|
3FH8
 
 | |
6X42
 
 | | High Resolution Crystal Structure Analysis of SERA5E from plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CALCIUM ION, ... | | Authors: | Clarke, O.B, Smith, N.A, Lee, M, Smith, B.J. | | Deposit date: | 2020-05-21 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the Plasmodium falciparum PfSERA5 pseudo-zymogen.
Protein Sci., 29, 2020
|
|
5OE2
 
 | | CRYSTAL STRUCTURE OF THE BETA-LACTAMASE OXA-245 | | Descriptor: | Beta-lactamase, CHLORIDE ION, SODIUM ION | | Authors: | Lund, B.A, Leiros, H.K.S. | | Deposit date: | 2017-07-07 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, activity and thermostability investigations of OXA-163, OXA-181 and OXA-245 using biochemical analysis, crystal structures and differential scanning calorimetry analysis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
