3VQA
 
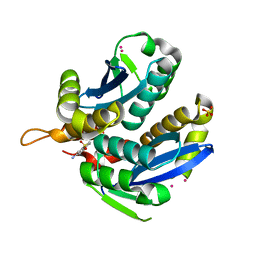 | | HIV-1 IN core domain in complex with 1-benzothiophen-6-amine 1,1-dioxide | | Descriptor: | 1-benzothiophen-6-amine 1,1-dioxide, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
2JIZ
 
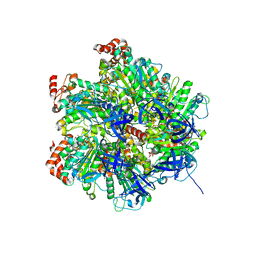 | | The Structure of F1-ATPase inhibited by resveratrol. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE GAMMA CHAIN, ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, ... | | Authors: | Gledhill, J.R, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Inhibition of Bovine F1-ATPase by Resveratrol and Related Polyphenols.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1NKE
 
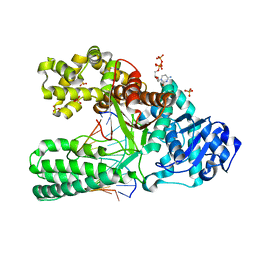 | | A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A CYTOSINE-THYMINE MISMATCH AFTER A SINGLE ROUND OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP. | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA POLYMERASE I, DNA PRIMER STRAND, ... | | Authors: | Johnson, S.J, Beese, L.S. | | Deposit date: | 2003-01-02 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of mismatch replication errors observed in a DNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
1NBX
 
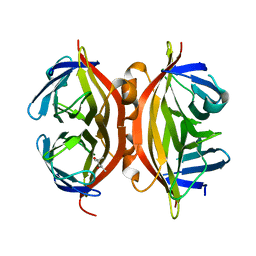 | | Streptavidin Mutant Y43A at 1.70A Resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-12-04 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3VXX
 
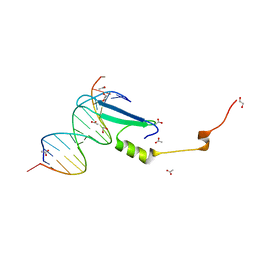 | | Crystal structure of methyl CpG binding domain of MBD4 in complex with the 5mCG/5mCG sequence | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*GP*TP*CP*(5CM)P*GP*GP*TP*AP*GP*TP*GP*AP*CP*T)-3'), ... | | Authors: | Otani, J, Arita, K, Kato, T, Kinoshita, M, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2012-09-21 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4
J.Biol.Chem., 288, 2013
|
|
2H2D
 
 | |
1NPB
 
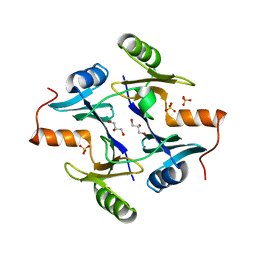 | | Crystal structure of the fosfomycin resistance protein from transposon Tn2921 | | Descriptor: | GLYCEROL, SULFATE ION, fosfomycin-resistance protein | | Authors: | Pakhomova, S, Rife, C.L, Armstrong, R.N, Newcomer, M.E. | | Deposit date: | 2003-01-17 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of fosfomycin resistance protein FosA from transposon Tn2921.
Protein Sci., 13, 2004
|
|
2HBK
 
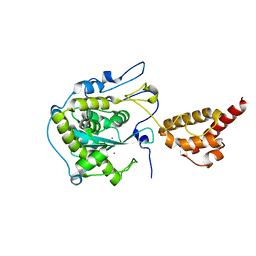 | | Structure of the yeast nuclear exosome component, Rrp6p, reveals an interplay between the active site and the HRDC domain; Protein in complex with Mn | | Descriptor: | Exosome complex exonuclease RRP6, MANGANESE (II) ION | | Authors: | Midtgaard, S.F, Assenholt, J, Jonstrup, A.T, Van, L.B, Jensen, T.H, Brodersen, D.E. | | Deposit date: | 2006-06-14 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1NT2
 
 | | CRYSTAL STRUCTURE OF FIBRILLARIN/NOP5P COMPLEX | | Descriptor: | Fibrillarin-like pre-rRNA processing protein, S-ADENOSYLMETHIONINE, conserved hypothetical protein | | Authors: | Aittaleb, M, Rashid, R, Chen, Q, Palmer, J.R, Daniels, C.J, Li, H. | | Deposit date: | 2003-01-28 | | Release date: | 2003-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and function of archaeal box C/D sRNP core proteins.
Nat.Struct.Biol., 10, 2003
|
|
3WK3
 
 | |
2GTF
 
 | | Crystal structure of nitrophorin 2 complex with pyrimidine | | Descriptor: | Nitrophorin-2, PROTOPORPHYRIN IX CONTAINING FE, PYRIMIDINE | | Authors: | Weichsel, A, Montfort, W.R. | | Deposit date: | 2006-04-27 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures, ligand induced conformational change and heme deformation in complexes of nitrophorin 2, a nitric oxide transport protein from rhodnius prolixus
To be Published
|
|
2GRE
 
 | |
3WK0
 
 | |
2JJ2
 
 | | The Structure of F1-ATPase inhibited by quercetin. | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE GAMMA CHAIN, ... | | Authors: | Gledhill, J.R, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Inhibition of Bovine F1-ATPase by Resveratrol and Related Polyphenols.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1QC6
 
 | | EVH1 domain from ENA/VASP-like protein in complex with ACTA peptide | | Descriptor: | EVH1 DOMAIN FROM ENA/VASP-LIKE PROTEIN, PHE-GLU-PHE-PRO-PRO-PRO-PRO-THR-ASP-GLU-GLU | | Authors: | Fedorov, A.A, Fedorov, E.V, Gertler, F.B, Almo, S.C. | | Deposit date: | 1999-05-17 | | Release date: | 1999-05-25 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of EVH1, a novel proline-rich ligand-binding module involved in cytoskeletal dynamics and neural function
Nat.Struct.Biol., 6, 1999
|
|
2G44
 
 | | Human Estrogen Receptor Alpha Ligand-Binding Domain In Complex With OBCP-1M-G and A Glucocorticoid Receptor Interacting Protein 1 NR Box II Peptide | | Descriptor: | 4-[(1S,2R,5S)-4,4,8-TRIMETHYL-3-OXABICYCLO[3.3.1]NON-7-EN-2-YL]PHENOL, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Rajan, S.S, Hsieh, R.W, Sharma, S.K, Greene, G.L. | | Deposit date: | 2006-02-21 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and characterization of novel estrogen
receptor agonist ligands and development of biochips for
nuclear receptor drug discovery
Thesis, 2006
|
|
1Q6Q
 
 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound xylitol 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-XYLITOL 5-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2003-08-13 | | Release date: | 2003-10-28 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Structural Evidence for a 1,2-Enediolate Intermediate in the Reaction Catalyzed by 3-Keto-l-Gulonate 6-Phosphate Decarboxylase, a Member of the Orotidine 5'-Monophosphate Decarboxylase Suprafamily
Biochemistry, 42, 2003
|
|
1Q93
 
 | | Crystal structure of a mutant of the sarcin/ricin domain from rat 28S rRNA | | Descriptor: | SODIUM ION, SULFATE ION, Sarcin/Ricin 28S rRNA | | Authors: | Correll, C.C, Beneken, J, Plantinga, M.J, Lubbers, M, Chan, Y.L. | | Deposit date: | 2003-08-22 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The common and distinctive features of the bulged-G motif based on a 1.04 A resolution RNA structure
Nucleic Acids Res., 31, 2003
|
|
2G8N
 
 | | Structure of hPNMT with inhibitor 3-Hydroxymethyl-7-(N-4-chlorophenylaminosulfonyl)-THIQ and AdoHcy | | Descriptor: | (3R)-N-(4-CHLOROPHENYL)-3-(HYDROXYMETHYL)-1,2,3,4-TETRAHYDROISOQUINOLINE-7-SULFONAMIDE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Gee, C.L, Martin, J.L. | | Deposit date: | 2006-03-02 | | Release date: | 2006-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Comparison of the Binding of 3-Fluoromethyl-7-sulfonyl-1,2,3,4-tetrahydroisoquinolines with Their Isosteric Sulfonamides to the Active Site of Phenylethanolamine N-Methyltransferase
J.Med.Chem., 49, 2006
|
|
1QBZ
 
 | | THE CRYSTAL STRUCTURE OF THE SIV GP41 ECTODOMAIN AT 1.47 A | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CHLORIDE ION, MERCURY (II) ION, ... | | Authors: | Yang, Z.-N, Mueser, T.C, Kaufman, J, Stahl, S.J, Wingfield, P.T, Hyde, C.C. | | Deposit date: | 1999-04-28 | | Release date: | 1999-05-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The crystal structure of the SIV gp41 ectodomain at 1.47 A resolution.
J.Struct.Biol., 126, 1999
|
|
2GEC
 
 | | Structure of the N-terminal domain of avian infectious bronchitis virus nucleocapsid protein (strain Gray) in a novel dimeric arrangement | | Descriptor: | Nucleocapsid protein | | Authors: | Jayaram, H, Fan, H, Bowman, B.R, Ooi, A, Jayaram, J, Collisson, E.W, Lescar, J, Prasad, B.V. | | Deposit date: | 2006-03-19 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation.
J.Virol., 80, 2006
|
|
1QVF
 
 | | Structure of a deacylated tRNA minihelix bound to the E site of the large ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S ribosomal rna, 50S RIBOSOMAL PROTEIN L10E, 50S ribosomal protein L13P, ... | | Authors: | Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2003-08-27 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit
RNA, 9, 2003
|
|
1QBF
 
 | |
2GA6
 
 | | The crystal structure of SARS nsp10 without zinc ion as additive | | Descriptor: | ZINC ION, orf1a polyprotein | | Authors: | Su, D, Lou, Z, Sun, F, Zhai, Y, Yang, H, Rao, Z. | | Deposit date: | 2006-03-08 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dodecamer Structure of Severe Acute Respiratory Syndrome Coronavirus Nonstructural Protein nsp10
J.Virol., 80, 2006
|
|
2JR7
 
 | | Solution structure of human DESR1 | | Descriptor: | DPH3 homolog, ZINC ION | | Authors: | Wu, F, Wu, J, Shi, Y. | | Deposit date: | 2007-06-21 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human DESR1, a CSL zinc-binding protein
Proteins, 71, 2008
|
|
