4MJ2
 
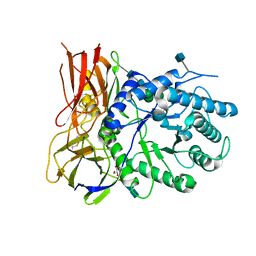 | | Crystal structure of apo-iduronidase in the R3 form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-L-iduronidase, ... | | Authors: | Bie, H, Yin, J, He, X, Kermode, A.R, Goddard-Borger, E.D, Withers, S.G, James, M.N.G. | | Deposit date: | 2013-09-03 | | Release date: | 2013-09-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into mucopolysaccharidosis I from the structure and action of alpha-L-iduronidase.
Nat.Chem.Biol., 9, 2013
|
|
4N95
 
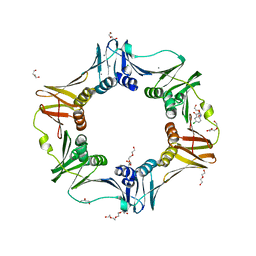 | | E. coli sliding clamp in complex with 5-chloroindoline-2,3-dione | | Descriptor: | 5-chloro-1H-indole-2,3-dione, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
5KGP
 
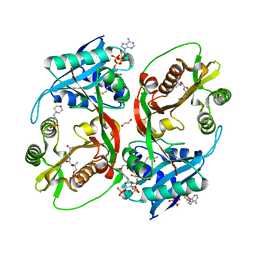 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum in complex with chitosan | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-alpha-D-glucopyranose, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
4N9I
 
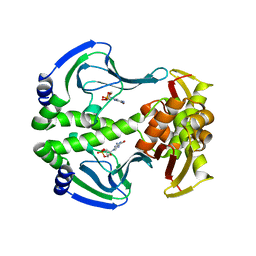 | | Crystal Structure of Transcription regulation protein CRP complexed with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Catabolite gene activator | | Authors: | Lee, B.-J, Seok, S.-H, Im, H, Yoon, H.-J. | | Deposit date: | 2013-10-21 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures of inactive CRP species reveal the atomic details of the allosteric transition that discriminates cyclic nucleotide second messengers.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5KH3
 
 | | Crystal structure of fragment (3-(5-Chloro-1,3-benzothiazol-2-yl)propanoic acid) bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(5-chloranyl-1,3-benzothiazol-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Dong, A, Ravichandran, M, Ferreira de Freitas, R, Schapira, M, Bountra, C, Edwards, A.M, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
4MJI
 
 | | T cell response to a HIV reverse transcriptase epitope presented by the protective allele HLA-B*51:01 | | Descriptor: | Beta-2-microglobulin, HIV Reverse Transcriptase peptide Marker, HLA class I histocompatibility antigen, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Sewell, A.K, Motozono, C, Takiguchi, M. | | Deposit date: | 2013-09-03 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Molecular basis of a dominant T cell response to an HIV reverse transcriptase 8-mer epitope presented by the protective allele HLA-B*51:01
J.Immunol., 192, 2014
|
|
5MIJ
 
 | | X-ray structure of carboplatin-encapsulated horse spleen apoferritin | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferritin light chain, ... | | Authors: | Pontillo, N, Ferraro, G, Helliwell, J.R, Merlino, A. | | Deposit date: | 2016-11-28 | | Release date: | 2017-03-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | X-ray Structure of the Carboplatin-Loaded Apo-Ferritin Nanocage.
ACS Med Chem Lett, 8, 2017
|
|
5U7K
 
 | | PDE2 catalytic domain complexed with inhibitors | | Descriptor: | 3-[5-(4-ethylphenyl)-1-methyl-1H-pyrazol-4-yl]-5-propoxy[1,2,4]triazolo[4,3-a]pyrazine, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Pandit, J, Parris, K. | | Deposit date: | 2016-12-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Application of Structure-Based Design and Parallel Chemistry to Identify a Potent, Selective, and Brain Penetrant Phosphodiesterase 2A Inhibitor.
J. Med. Chem., 60, 2017
|
|
6JQZ
 
 | | ZHD/H242A complex with ZEN | | Descriptor: | (3S,11E)-14,16-dihydroxy-3-methyl-3,4,5,6,9,10-hexahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, GLYCEROL, Zearalenone hydrolase | | Authors: | Hu, X.J. | | Deposit date: | 2019-04-02 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of ZHD complex
To Be Published
|
|
6JR6
 
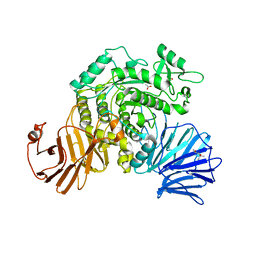 | | Flavobacterium johnsoniae GH31 dextranase, FjDex31A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Candidate alpha-glycosidase Glycoside hydrolase family 31 | | Authors: | Tonozuka, T. | | Deposit date: | 2019-04-02 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into polysaccharide recognition by Flavobacterium johnsoniae dextranase, a member of glycoside hydrolase family 31.
Febs J., 287, 2020
|
|
4MOZ
 
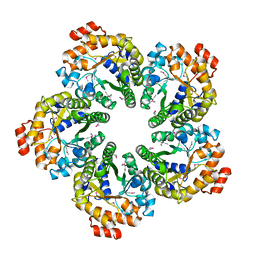 | |
5MDZ
 
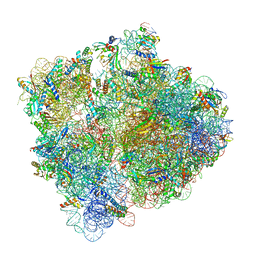 | | Structure of the 70S ribosome (empty A site) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | James, N.R, Brown, A, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-11-13 | | Release date: | 2016-12-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Translational termination without a stop codon.
Science, 354, 2016
|
|
2R3F
 
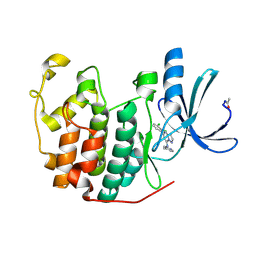 | | Crystal Structure of Cyclin-Dependent Kinase 2 with inhibitor | | Descriptor: | 5-(2,3-dichlorophenyl)-N-(pyridin-4-ylmethyl)pyrazolo[1,5-a]pyrimidin-7-amine, Cell division protein kinase 2 | | Authors: | Fischmann, T.O, Hruza, A.W, Madison, V.M, Duca, J.S. | | Deposit date: | 2007-08-29 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided discovery of cyclin-dependent kinase inhibitors.
Biopolymers, 89, 2008
|
|
4MQ6
 
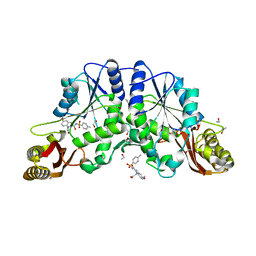 | |
4MQP
 
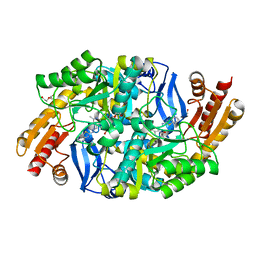 | | Mycobaterium tuberculosis transaminase BioA complexed with 2-hydrazinylbenzo[d]thiazole | | Descriptor: | (4-{[(E)-1,3-benzothiazol-2-yldiazenyl]methyl}-5-hydroxy-6-methylpyridin-3-yl)methyl dihydrogen phosphate, 1,2-ETHANEDIOL, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, ... | | Authors: | Finzel, B.C, Dai, R. | | Deposit date: | 2013-09-16 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Inhibition of Mycobacterium tuberculosis Transaminase BioA by Aryl Hydrazines and Hydrazides.
Chembiochem, 15, 2014
|
|
5JR9
 
 | | Crystal structure of apo-NeC3PO | | Descriptor: | NEQ131 | | Authors: | Zhang, J, Gan, J. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for single-stranded RNA recognition and cleavage by C3PO
Nucleic Acids Res., 44, 2016
|
|
4MUM
 
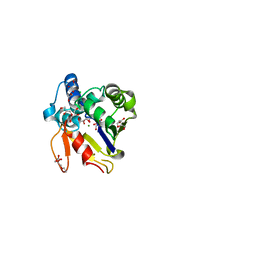 | |
4MOR
 
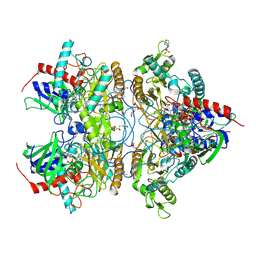 | | Pyranose 2-oxidase H450G/V546C double mutant with 3-fluorinated galactose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-deoxy-3-fluoro-beta-D-galactopyranose, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Spadiut, O, Gandini, R, Haltrich, D, Divne, C. | | Deposit date: | 2013-09-12 | | Release date: | 2014-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Binding of Fluorinated Glucose and Galactose to Trametes multicolor Pyranose 2-Oxidase Variants with Improved Galactose Conversion.
Plos One, 9, 2014
|
|
1EA8
 
 | |
4MR4
 
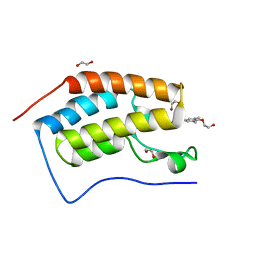 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolinone ligand (RVX-208) | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4MWD
 
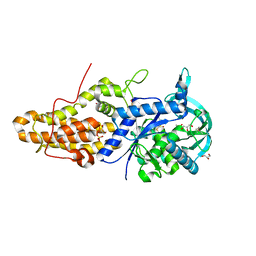 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-thiophen-3-ylurea (Chem 1433) and ATP analog AMPPCP | | Descriptor: | 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4MX7
 
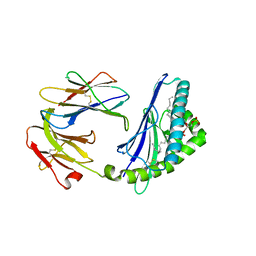 | | Structure of mouse CD1d in complex with dioleoyl-phosphatidic acid | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl (9Z,9'Z)bis-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2013-09-25 | | Release date: | 2014-10-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | TBN
To be Published
|
|
4MYY
 
 | |
5U36
 
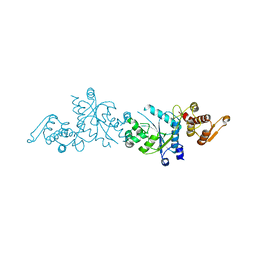 | | Crystal Structure Of A Mutant M. Jannashii Tyrosyl-tRNA Synthetase | | Descriptor: | Tyrosine--tRNA ligase | | Authors: | Luo, X, Fu, G, Zhu, X, Wilson, I.A, Wang, F. | | Deposit date: | 2016-12-01 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Genetically encoding phosphotyrosine and its nonhydrolyzable analog in bacteria.
Nat. Chem. Biol., 13, 2017
|
|
5K2F
 
 | | Structure of NNQQNY from yeast prion Sup35 with cadmium acetate determined by MicroED | | Descriptor: | ACETATE ION, CADMIUM ION, Eukaryotic peptide chain release factor GTP-binding subunit | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
