9C3H
 
 | | Structure of the CNOT3-bound human 80S ribosome with tRNA-ARG in the P-site. | | Descriptor: | 18S rRNA, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 28S rRNA, ... | | Authors: | Erzberger, J.P, Cruz, V.E. | | Deposit date: | 2024-06-01 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Specific tRNAs promote mRNA decay by recruiting the CCR4-NOT complex to translating ribosomes.
Science, 386, 2024
|
|
7RFP
 
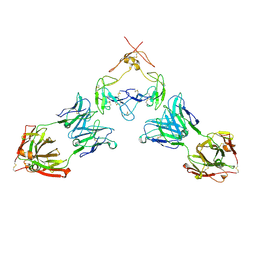 | | Mouse GITR (mGITR) with DTA-1 Fab fragment | | Descriptor: | DTA-1 (heavy chain), DTA-1 (light chain), Tumor necrosis factor receptor superfamily member 18,Enhanced green fluorescent protein | | Authors: | Meyerson, J.R, He, C. | | Deposit date: | 2021-07-14 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Therapeutic antibody activation of the glucocorticoid-induced TNF receptor by a clustering mechanism.
Sci Adv, 8, 2022
|
|
7Q0F
 
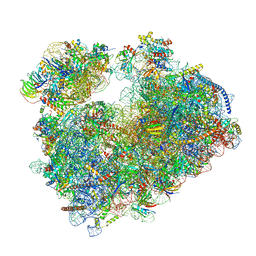 | | Structure of Candida albicans 80S ribosome in complex with phyllanthoside | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Zgadzay, Y, Kolosova, O, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-14 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
7Q08
 
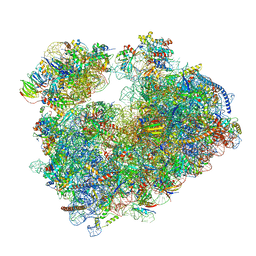 | | Structure of Candida albicans 80S ribosome in complex with cycloheximide | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Zgadzay, Y, Kolosova, O, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-14 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
7PWF
 
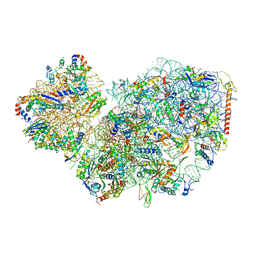 | | Cryo-EM structure of small subunit of Giardia lamblia ribosome at 2.9 A resolution | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S25, 40S ribosomal protein S26, ... | | Authors: | Hiregange, D.G, Rivalta, A, Bose, T, Breiner-Goldstein, E, Samiya, S, Cimicata, G, Kulakova, L, Zimmerman, E, Bashan, A, Herzberg, O, Yonath, A. | | Deposit date: | 2021-10-06 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure of the ancient eukaryotic ribosome from the human parasite Giardia lamblia.
Nucleic Acids Res., 50, 2022
|
|
7PZY
 
 | | Structure of the vacant Candida albicans 80S ribosome | | Descriptor: | 1,4-DIAMINOBUTANE, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Zgadzay, Y, Kolosova, O, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-13 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
7QIX
 
 | | Specific features and methylation sites of a plant ribosome. 40S body ribosomal subunit. | | Descriptor: | 18S rRNA body, 30S ribosomal protein S15, chloroplastic, ... | | Authors: | Cottilli, P, Itoh, Y, Amunts, A. | | Deposit date: | 2021-12-16 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Cryo-EM structure and rRNA modification sites of a plant ribosome.
Plant Commun., 3, 2022
|
|
7Q0R
 
 | | Structure of the Candida albicans 80S ribosome in complex with blasticidin s | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Kolosova, O, Zgadzay, Y, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-16 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
6QEV
 
 | | EngBF DARPin Fusion 4b B6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MANGANESE (II) ION, PEGA domain-containing protein,PEGA domain-containing protein,EngBF DARPin fusion B6 complex, ... | | Authors: | Ernst, P, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2019-01-08 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of biological targets by host:guest crystal lattice engineering.
Sci Rep, 9, 2019
|
|
6QFK
 
 | | EngBF DARPin Fusion 4b G10 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Ernst, P, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of biological targets by host:guest crystal lattice engineering.
Sci Rep, 9, 2019
|
|
4AKE
 
 | | ADENYLATE KINASE | | Descriptor: | ADENYLATE KINASE | | Authors: | Schlauderer, G.J, Schulz, G.E. | | Deposit date: | 1995-12-29 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Adenylate kinase motions during catalysis: an energetic counterweight balancing substrate binding.
Structure, 4, 1996
|
|
4QBF
 
 | | Crystal structure of a stable adenylate kinase variant AKlse2 | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ZINC ION | | Authors: | Moon, S, Bae, E. | | Deposit date: | 2014-05-08 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effectiveness and limitations of local structural entropy optimization in the thermal stabilization of mesophilic and thermophilic adenylate kinases.
Proteins, 82, 2014
|
|
7QGG
 
 | | Neuronal RNA granules are ribosome complexes stalled at the pre-translocation state | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Pulk, A, Kipper, K, Mansour, A. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Neuronal RNA granules are ribosome complexes stalled at the pre-translocation state.
J.Mol.Biol., 434, 2022
|
|
6OHB
 
 | | E. coli Guanine Deaminase | | Descriptor: | Guanine deaminase, ZINC ION | | Authors: | Shek, R.S, French, J.B. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Determinants for Substrate Selectivity in Guanine Deaminase Enzymes of the Amidohydrolase Superfamily.
Biochemistry, 58, 2019
|
|
7QIZ
 
 | | Specific features and methylation sites of a plant 80S ribosome | | Descriptor: | 1,4-DIAMINOBUTANE, 18S, 25S rRNA, ... | | Authors: | Cottilli, P, Itoh, Y, Amunts, A. | | Deposit date: | 2021-12-16 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM structure and rRNA modification sites of a plant ribosome.
Plant Commun., 3, 2022
|
|
6MTC
 
 | | Rabbit 80S ribosome with Z-site tRNA and IFRD2 (unrotated state) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | Deposit date: | 2018-10-19 | | Release date: | 2018-11-21 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
6P5J
 
 | | Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 2) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
2Z6Q
 
 | |
6MTE
 
 | | Rabbit 80S ribosome with eEF2 and SERBP1 (rotated state) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | Deposit date: | 2018-10-19 | | Release date: | 2018-11-21 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
6Q8Y
 
 | | Cryo-EM structure of the mRNA translating and degrading yeast 80S ribosome-Xrn1 nuclease complex | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Heckel, E, Cheng, J, Buschauer, R, Kater, L, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2018-12-16 | | Release date: | 2019-03-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the 80S ribosome-Xrn1 nuclease complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7QP6
 
 | | Structure of the human 48S initiation complex in open state (h48S AUG open) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Yi, S.-H, Petrychenko, V, Schliep, J.E, Goyal, A, Linden, A, Chari, A, Urlaub, H, Stark, H, Rodnina, M.V, Adio, S, Fischer, N. | | Deposit date: | 2022-01-03 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Conformational rearrangements upon start codon recognition in human 48S translation initiation complex.
Nucleic Acids Res., 50, 2022
|
|
7QP7
 
 | | Structure of the human 48S initiation complex in closed state (h48S AUG closed) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Yi, S.-H, Petrychenko, V, Schliep, J.E, Goyal, A, Linden, A, Chari, A, Urlaub, H, Stark, H, Rodnina, M.V, Adio, S, Fischer, N. | | Deposit date: | 2022-01-03 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Conformational rearrangements upon start codon recognition in human 48S translation initiation complex.
Nucleic Acids Res., 50, 2022
|
|
4AU5
 
 | | Structure of the NhaA dimer, crystallised at low pH | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, NA(+)/H(+) ANTIPORTER NHAA, SULFATE ION | | Authors: | Drew, D, Lee, C, Iwata, S, Cameron, A.D. | | Deposit date: | 2012-05-14 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.696 Å) | | Cite: | Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights.
J. Gen. Physiol., 144, 2014
|
|
6OLG
 
 | |
4CF7
 
 | | Crystal structure of adenylate kinase from Aquifex aeolicus with MgADP bound | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENYLATE KINASE, ... | | Authors: | Kerns, S.J, Agafonov, R.V, Cho, Y.-J, Pontiggia, F, Otten, R, Pachov, D.V, Kutter, S, Phung, L.A, Murphy, P.N, Thai, V, Hagan, M.F, Kern, D. | | Deposit date: | 2013-11-13 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | The Energy Landscape of Adenylate Kinase During Catalysis.
Nat.Struct.Mol.Biol., 22, 2015
|
|
