9F9L
 
 | | Crystal structure of MUS81-EME1 bound by compound 16. | | Descriptor: | 2-[2-[4-(cyanomethyl)phenyl]phenyl]-5-oxidanyl-6-oxidanylidene-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9F98
 
 | | Crystal structure of MUS81-EME1, apo form. | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81 | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9F8G
 
 | |
9BOX
 
 | | Room-temperature X-ray structure of human mitochondrial serine hydroxymethyltransferase (hSHMT2) with PLP-glycine external aldimine and 5-formyltetrahydrofolate (folinic acid) | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)glycine, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, Serine hydroxymethyltransferase, ... | | Authors: | Drago, V.N, Kovalevsky, A. | | Deposit date: | 2024-05-06 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Universality of critical active site glutamate as an acid-base catalyst in serine hydroxymethyltransferase function.
Chem Sci, 15, 2024
|
|
9BOW
 
 | | X-ray structure of Thermus thermophilus serine hydroxymethyltransferase with PLP-L-Ser external aldimine and 5-formyltetrahydrofolate (folinic acid) | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SERINE, ... | | Authors: | Drago, V.N, Kovalevsky, A. | | Deposit date: | 2024-05-06 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Universality of critical active site glutamate as an acid-base catalyst in serine hydroxymethyltransferase function.
Chem Sci, 15, 2024
|
|
9BOR
 
 | | IkappaBzeta ankyrin repeat domain:NF-kappaB p50 homodimer complex at 2.0 Angstrom resolution | | Descriptor: | (2R,3S)-heptane-1,2,3-triol, NF-kappa-B inhibitor zeta, Nuclear factor NF-kappa-B p50 subunit | | Authors: | Rogers, W.E, Zhu, N, Huxford, T. | | Deposit date: | 2024-05-05 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural and biochemical analyses of the nuclear I kappa B zeta protein in complex with the NF-kappa B p50 homodimer.
Genes Dev., 38, 2024
|
|
9BOQ
 
 | | Human p97/VCP structure with a triazole inhibitor (NSC799462/dodecamer) | | Descriptor: | 3-(2,6-difluoro-4-{[(4P)-5-{[(2S)-hexan-2-yl]sulfanyl}-4-(pyridin-3-yl)-4H-1,2,4-triazol-3-yl]methoxy}phenyl)prop-2-yn-1-yl (1-methylpiperidin-4-yl)carbamate, ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, DeVore, K, Chiu, P.-L. | | Deposit date: | 2024-05-05 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Mechanism of allosteric inhibition of human p97/VCP ATPase and its disease mutant by triazole inhibitors.
Commun Chem, 7, 2024
|
|
9BOH
 
 | | Room-temperature X-ray structure of Thermus Thermophilus serine hydroxymethyltransferase (SHMT) with PLP-glycine external aldimine and 5-formyltetrahydrofolate (folinic acid) | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)glycine, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SULFATE ION, ... | | Authors: | Drago, V.N, Kovalevsky, A. | | Deposit date: | 2024-05-03 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Universality of critical active site glutamate as an acid-base catalyst in serine hydroxymethyltransferase function.
Chem Sci, 15, 2024
|
|
9F7H
 
 | | UP1 in complex with Z45617795 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, N-(2-phenylethyl)methanesulfonamide | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-05-03 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F7F
 
 | | UP1 in complex with Z1491353358 | | Descriptor: | (3S)-1-(phenylsulfonyl)pyrrolidin-3-amine, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-05-03 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9BOL
 
 | | Crystal structure of the complex between VHL, ElonginB, ElonginC, and compound 5 | | Descriptor: | (4R)-1-[(2S)-2-(4-cyclopropyl-1H-1,2,3-triazol-1-yl)-3,3-dimethylbutanoyl]-4-hydroxy-N-{[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl}-L-prolinamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Murray, J.M, Wu, H, Fuhrmann, J, Fairbrother, W.J, DiPasquale, A. | | Deposit date: | 2024-05-03 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Potency-enhanced peptidomimetic VHL ligands with improved oral bioavailability
To Be Published
|
|
9BNR
 
 | | 4-(2-isothiocyanatoethyl)benzenesulfonamide complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | Macrophage migration inhibitory factor, N-[2-(4-sulfamoylphenyl)ethyl]methanethioamide, SULFATE ION | | Authors: | Fellner, M, Rutledge, M.T, Putha, L, Kok, L.K, Gamble, A.B, Wilbanks, S.M, Vernall, A.J, Tyndall, J.D.A. | | Deposit date: | 2024-05-02 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Covalent isothiocyanate inhibitors of macrophage migration inhibitory factor as potential colorectal cancer treatments.
Chemmedchem, 2024
|
|
9BNQ
 
 | | N-(4-(isothiocyanatomethyl)phenyl)methanesulfonamide complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, N-{[4-(methanesulfonamido)phenyl]methyl}methanethioamide, ... | | Authors: | Fellner, M, Rutledge, M.T, Putha, L, Kok, L.K, Gamble, A.B, Wilbanks, S.M, Vernall, A.J, Tyndall, J.D.A. | | Deposit date: | 2024-05-02 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Covalent isothiocyanate inhibitors of macrophage migration inhibitory factor as potential colorectal cancer treatments.
Chemmedchem, 2024
|
|
9F6I
 
 | |
9F6D
 
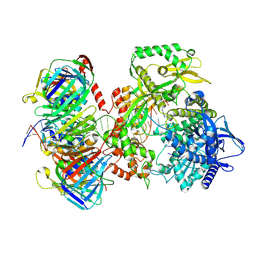 | | Human DNA polymerase epsilon bound to DNA and PCNA (open conformation) | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, DNA nascent strand, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Roske, J.J, Yeeles, J.T.P. | | Deposit date: | 2024-05-01 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for processive daughter-strand synthesis and proofreading by the human leading-strand DNA polymerase Pol epsilon.
Nat.Struct.Mol.Biol., 2024
|
|
9F6L
 
 | |
9F6J
 
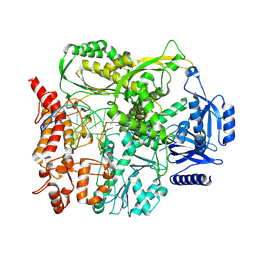 | |
9F6F
 
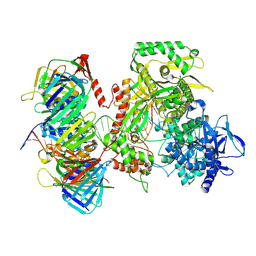 | | Human DNA polymerase epsilon bound to DNA and PCNA (closed conformation) | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, DNA nascent strand, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Roske, J.J, Yeeles, J.T.P. | | Deposit date: | 2024-05-01 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural basis for processive daughter-strand synthesis and proofreading by the human leading-strand DNA polymerase Pol epsilon.
Nat.Struct.Mol.Biol., 2024
|
|
9F6K
 
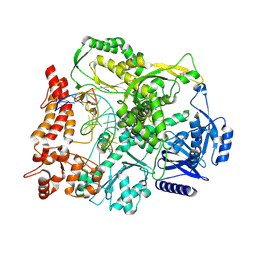 | |
9F6E
 
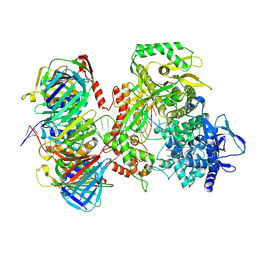 | | Human DNA polymerase epsilon bound to DNA and PCNA (ajar conformation) | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, DNA nascent strand, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Roske, J.J, Yeeles, J.T.P. | | Deposit date: | 2024-05-01 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural basis for processive daughter-strand synthesis and proofreading by the human leading-strand DNA polymerase Pol epsilon.
Nat.Struct.Mol.Biol., 2024
|
|
9F5S
 
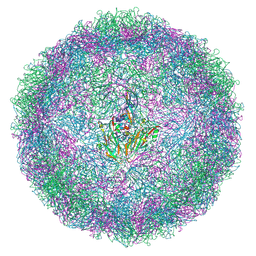 | | EVA71 E096A native particle | | Descriptor: | SPHINGOSINE, VP0, VP1, ... | | Authors: | Kingston, N.J, Stonehouse, N.J, Rowlands, D.J, Hogle, J.M, Filman, D.J, Snowden, J.S.S. | | Deposit date: | 2024-04-30 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Reappraisal of the mechanism of the enterovirus VP0 maturation cleavage based on the structure of a stabilised assembly intermediate
To Be Published
|
|
9BLN
 
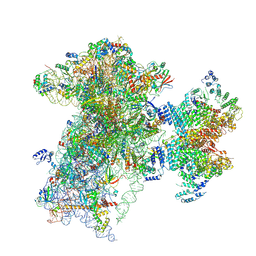 | | The structure of human Pdcd4 bound to the 40S-eIF4A-eIF3-eIF1 complex | | Descriptor: | 40S ribosomal protein S14, ACETIC ACID, Eukaryotic initiation factor 4A-I, ... | | Authors: | Brito Querido, J, Sokabe, M, Diaz-Lopez, I, Gordiyenko, Y, Zuber, P, Yifei, D, Albacete-Albacete, L, Ramakrishnan, V, S Fraser, C. | | Deposit date: | 2024-04-30 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Human tumor suppressor protein Pdcd4 binds at the mRNA entry channel in the 40S small ribosomal subunit.
Nat Commun, 15, 2024
|
|
9F6A
 
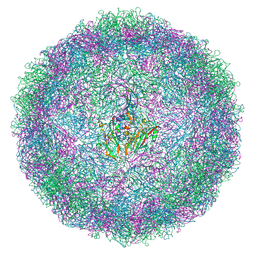 | | EVA71 E096A native particle | | Descriptor: | SPHINGOSINE, VP0, VP1, ... | | Authors: | Kingston, N.J, Stonehouse, N.J, Rowlands, D.J, Hogle, J.M, Filman, D.J, Snowden, J.S.S. | | Deposit date: | 2024-04-30 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Reappraisal of the mechanism of the enterovirus VP0 maturation cleavage based on the structure of a stabilised assembly intermediate
To Be Published
|
|
9F5K
 
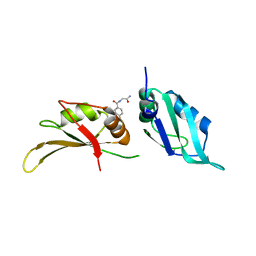 | | UP1 in complex with Z33546965 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, ~{N}-(2-azanyl-2-oxidanylidene-ethyl)-4-methoxy-benzamide | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-29 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9BKJ
 
 | | Cholecystokinin 1 receptor (CCK1R) Y140A mutant, Gq chimera (mGsqi) complex | | Descriptor: | AMINO GROUP, Cholecystokinin receptor type A, Cholecystokinin-8, ... | | Authors: | Cary, B.P, Harikumar, K.G, Zhao, P, Desai, A.J, Mobbs, J.M, Toufaily, C, Furness, S.G.B, Christopoulos, A, Belousoff, M.J, Wootten, D, Sexton, P.M, Miller, L.J. | | Deposit date: | 2024-04-29 | | Release date: | 2024-05-22 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Cholesterol-dependent dynamic changes in the conformation of the type 1 cholecystokinin receptor affect ligand binding and G protein coupling.
Plos Biol., 22, 2024
|
|
