2HCV
 
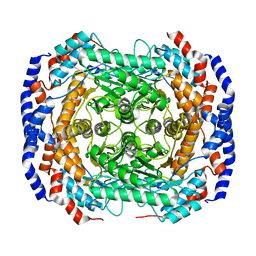 | | Crystal structure of L-rhamnose isomerase from Pseudomonas stutzeri with metal ion | | Descriptor: | L-rhamnose isomerase, ZINC ION | | Authors: | Yoshida, H, Yamada, M, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2006-06-19 | | Release date: | 2006-12-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structures of l-Rhamnose Isomerase from Pseudomonas stutzeri in Complexes with l-Rhamnose and d-Allose Provide Insights into Broad Substrate Specificity
J.Mol.Biol., 365, 2007
|
|
2Q09
 
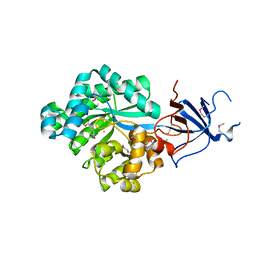 | | Crystal structure of Imidazolonepropionase from environmental sample with bound inhibitor 3-(2,5-Dioxo-imidazolidin-4-yl)-propionic acid | | Descriptor: | 3-[(4S)-2,5-DIOXOIMIDAZOLIDIN-4-YL]PROPANOIC ACID, FE (III) ION, Imidazolonepropionase | | Authors: | Tyagi, R, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-21 | | Release date: | 2007-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A common catalytic mechanism for proteins of the HutI family.
Biochemistry, 47, 2008
|
|
2XMN
 
 | | High resolution snapshots of defined TolC open states present an iris- like movement of periplasmic entrance helices | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Outer membrane protein TolC | | Authors: | Pei, X.Y, Koronakis, E, Hughes, C, Koronakis, V. | | Deposit date: | 2010-07-28 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of sequential open states in a symmetrical opening transition of the TolC exit duct.
Proc. Natl. Acad. Sci. U.S.A., 108, 2011
|
|
2Y48
 
 | | Crystal structure of LSD1-CoREST in complex with a N-terminal SNAIL peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC DEMETHYLASE 1A, REST COREPRESSOR 1, ... | | Authors: | Baron, R, Binda, C, Tortorici, M, McCammon, J.A, Mattevi, A. | | Deposit date: | 2011-01-05 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular Mimicry and Ligand Recognition in Binding and Catalysis by the Histone Demethylase Lsd1-Corest Complex.
Structure, 19, 2011
|
|
2Q51
 
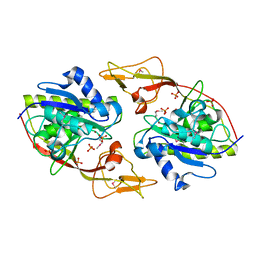 | | Ensemble refinement of the protein crystal structure of an aspartoacylase from Homo sapiens | | Descriptor: | Aspartoacylase, PHOSPHATE ION, ZINC ION | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2Q4Z
 
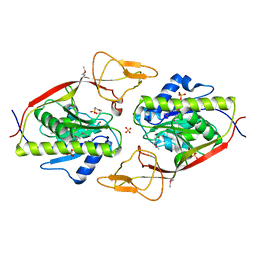 | | Ensemble refinement of the protein crystal structure of an aspartoacylase from Rattus norvegicus | | Descriptor: | Aspartoacylase, SULFATE ION, ZINC ION | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2L5N
 
 | | NMR Structure of YbbR family protein Dhaf_0833 (residues 32-118) from Desulfitobacterium hafniense DCB-2: Northeast Structural Genomics Consortium target DhR29B | | Descriptor: | YbbR family protein | | Authors: | Cort, J.R, Barb, A.W, Lee, H, Ramelot, T.A, Yang, Y, Belote, R.L, Ciccosanti, C.R, Haleema, J, Acton, T.B, Xiao, R.R, Everett, J.K, Montelione, G.T, Prestegard, J.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-11-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of domains I and IV from YbbR are representative of a widely distributed protein family.
Protein Sci., 20, 2011
|
|
2OHO
 
 | |
2L3U
 
 | | Solution Structure of Domain IV from the YbbR family protein of Desulfitobacterium hafniense: Northeast Structural Genomics Consortium target DhR29A | | Descriptor: | YbbR family protein | | Authors: | Barb, A.W, Lee, H, Belote, R.L, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-23 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of domains I and IV from YbbR are representative of a widely distributed protein family.
Protein Sci., 20, 2011
|
|
3KTU
 
 | |
3LT5
 
 | | X-ray Crystallographic structure of a Pseudomonas Aeruginosa Azoreductase in complex with balsalazide | | Descriptor: | (3E)-3-({4-[(2-carboxyethyl)carbamoyl]phenyl}hydrazono)-6-oxocyclohexa-1,4-diene-1-carboxylic acid, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, ... | | Authors: | Ryan, A, Laurieri, N, Westwood, I, Wang, C.-J, Lowe, E, Sim, E. | | Deposit date: | 2010-02-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Novel Mechanism for Azoreduction
J.Mol.Biol., 400, 2010
|
|
6J31
 
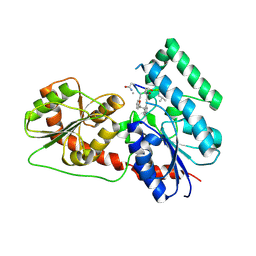 | | Crystal Structure Analysis of the Glycotransferase of kitacinnamycin | | Descriptor: | (2E,2'E)-3,3'-(1,2-phenylene)di(prop-2-enoic acid), DBB-DSG-VAL-MEA-VAL-GLY-GLY-DVA-DLE, kcn28 | | Authors: | Shi, J, Liu, C.L, Zhang, B, Guo, W.J, Zhu, J.P, Xu, X, Xu, Q, Jiao, R.H, Tan, R.X, Ge, H.M. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Genome mining and biosynthesis of kitacinnamycins as a STING activator.
Chem Sci, 10, 2019
|
|
3LNP
 
 | | Crystal Structure of Amidohydrolase family Protein OLEI01672_1_465 from Oleispira antarctica | | Descriptor: | ACETIC ACID, Amidohydrolase family Protein OLEI01672_1_465, CALCIUM ION, ... | | Authors: | Kim, Y, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-02 | | Release date: | 2010-02-16 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
2P8C
 
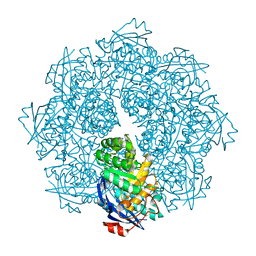 | | Crystal structure of N-succinyl Arg/Lys racemase from Bacillus cereus ATCC 14579 complexed with N-succinyl Arg. | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme family protein, N~2~-(3-CARBOXYPROPANOYL)-L-ARGININE | | Authors: | Fedorov, A.A, Song, L, Fedorov, E.V, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-03-22 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction and assignment of function for a divergent N-succinyl amino acid racemase.
Nat.Chem.Biol., 3, 2007
|
|
2LQL
 
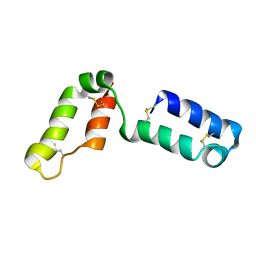 | | Solution structure of CHCH5 | | Descriptor: | Coiled-coil-helix-coiled-coil-helix domain-containing protein 5 | | Authors: | Peruzzini, R, Ciofi-Baffoni, S, Banci, L, Bertini, I. | | Deposit date: | 2012-03-09 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-06 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of CHCHD5 and CHCHD7: Two atypical human twin CX(9)C proteins.
J.Struct.Biol., 180, 2012
|
|
2P88
 
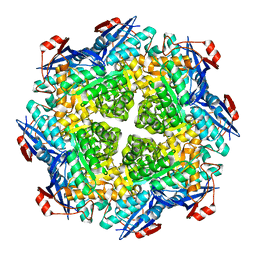 | | Crystal structure of N-succinyl Arg/Lys racemase from Bacillus cereus ATCC 14579 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme family protein | | Authors: | Fedorov, A.A, Song, L, Fedorov, E.V, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-03-22 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Prediction and assignment of function for a divergent N-succinyl amino acid racemase.
Nat.Chem.Biol., 3, 2007
|
|
2QT3
 
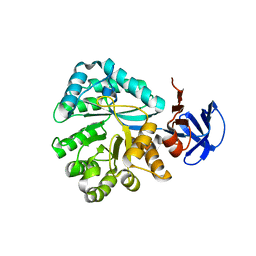 | | Crystal structure of N-Isopropylammelide isopropylaminohydrolase AtzC from Pseudomonas sp. strain ADP complexed with Zn | | Descriptor: | N-isopropylammelide isopropyl amidohydrolase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Seffernick, J, Wackett, L.P, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-01 | | Release date: | 2007-09-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structure of N-Isopropylammelide isopropylaminohydrolase AtzC from Pseudomonas sp. strain ADP complexed with Zn.
To be Published
|
|
2LQT
 
 | |
2OOF
 
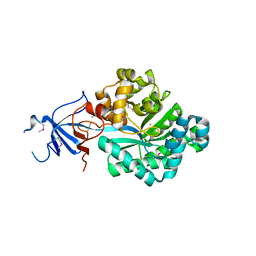 | |
2HPM
 
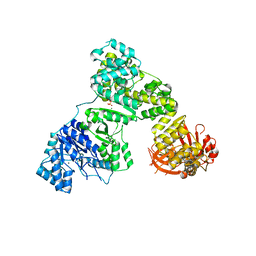 | | Eubacterial and Eukaryotic Replicative DNA Polymerases are not Homologous: X-ray Structure of DNA Polymerase III | | Descriptor: | CHLORIDE ION, DNA Polymerase III alpha subunit, MAGNESIUM ION, ... | | Authors: | Bailey, S, Wing, R.A, Steitz, T.A. | | Deposit date: | 2006-07-17 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The Structure of T. aquaticus DNA Polymerase III Is Distinct from Eukaryotic Replicative DNA Polymerases.
Cell(Cambridge,Mass.), 126, 2006
|
|
2LQ8
 
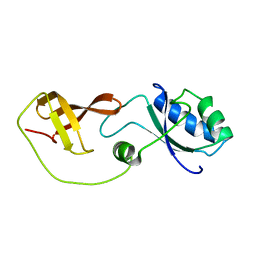 | | Domain interaction in Thermotoga maritima NusG | | Descriptor: | Transcription antitermination protein nusG | | Authors: | Droegemueller, J, Stegmann, C, Burmann, B, Roesch, P, Wahl, M.C, Schweimer, K. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An Autoinhibited State in the Structure of Thermotoga maritima NusG.
Structure, 21, 2013
|
|
2NOF
 
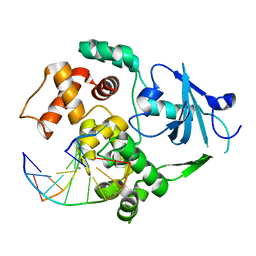 | | Structure of Q315F human 8-oxoguanine glycosylase proximal crosslink to 8-oxoguanine DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*C*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOI
 
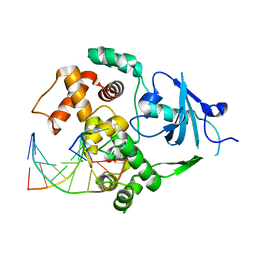 | | Structure of G42A human 8-oxoguanine glycosylase crosslinked to undamaged G-containing DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*C*CP*AP*GP*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOB
 
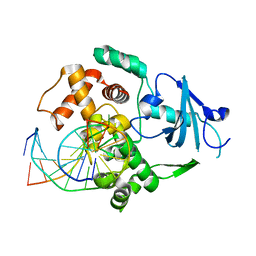 | | Structure of catalytically inactive H270A human 8-oxoguanine glycosylase crosslinked to 8-oxoguanine DNA | | Descriptor: | 5'-D(*G*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*T*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOE
 
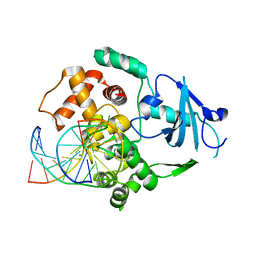 | | Structure of catalytically inactive G42A human 8-oxoguanine glycosylase complexed to 8-oxoguanine DNA | | Descriptor: | 5'-D(*G*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*G*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
