4YUP
 
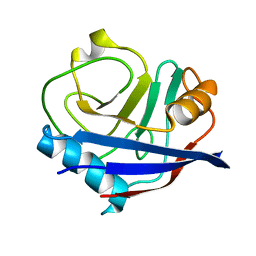 | | Multiconformer fixed-target X-ray free electron (XFEL) model of CypA at 273 K | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | Deposit date: | 2015-03-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
1T73
 
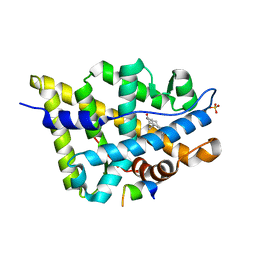 | | Crystal structure of the androgen receptor ligand binding domain in complex with a FxxFF motif | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, FxxFF motif peptide, ... | | Authors: | Hur, E, Pfaff, S.J, Payne, E.S, Gron, H, Buehrer, B.M, Fletterick, R.J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition and accommodation at the androgen receptor coactivator binding interface.
Plos Biol., 2, 2004
|
|
8V7R
 
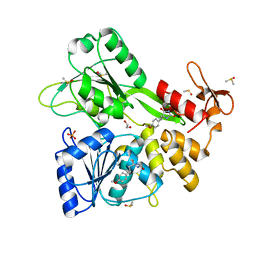 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56772132 | | Descriptor: | (5R)-5-[2-(4-methoxyphenyl)ethyl]-5-methylimidazolidine-2,4-dione, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56772132
To Be Published
|
|
8EFS
 
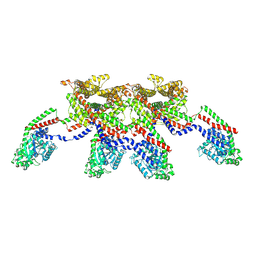 | | CryoEM of the soluble OPA1 tetramer from the apo helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1 | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-09 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (9.68 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
5GLJ
 
 | |
8EF7
 
 | | CryoEM of the soluble OPA1 dimer from the apo helical assembly on a lipid membrane | | Descriptor: | Dynamin-like 120 kDa protein, form S1 | | Authors: | Nyenhuis, S.B, Wu, X, Stanton, A.E, Strub, M.P, Yim, Y.I, Canagarajah, B, Hinshaw, J.E. | | Deposit date: | 2022-09-08 | | Release date: | 2023-06-28 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (9.68 Å) | | Cite: | OPA1 helical structures give perspective to mitochondrial dysfunction.
Nature, 620, 2023
|
|
5MRQ
 
 | | Arabidopsis thaliana IspD Asp262Ala Mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, ... | | Authors: | Schwab, A, Illarionov, B, Frank, A, Kunfermann, A, Seet, M, Bacher, A, Witschel, M, Fischer, M, Groll, M, Diederich, F. | | Deposit date: | 2016-12-23 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Allosteric Inhibition of the Enzyme IspD by Three Different Classes of Ligands.
ACS Chem. Biol., 12, 2017
|
|
1T22
 
 | | Structural basis for degenerate recognition of HIV peptide variants by cytotoxic lymphocyte, variant SL9, orthorhombic crystal | | Descriptor: | Beta-2-microglobulin, GAG PEPTIDE, HLA class I histocompatibility antigen, ... | | Authors: | Martinez-Hackert, E, Anikeeva, N, Kalams, S.A, Walker, B.D, Hendrickson, W.A, Sykulev, Y. | | Deposit date: | 2004-04-19 | | Release date: | 2005-09-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Degenerate Recognition of Natural HIV Peptide Variants by Cytotoxic Lymphocytes.
J.Biol.Chem., 281, 2006
|
|
4IYT
 
 | | Structure Of The Y184A Mutant Of The PANTON-VALENTINE LEUCOCIDIN S Component From STAPHYLOCOCCUS AUREUS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LukS-PV | | Authors: | Guerin, F, Laventie, B.J, Prevost, G, Mourey, L, Maveyraud, L. | | Deposit date: | 2013-01-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Residues essential for panton-valentine leukocidin s component binding to its cell receptor suggest both plasticity and adaptability in its interaction surface
Plos One, 9, 2014
|
|
1T2E
 
 | | Plasmodium falciparum lactate dehydrogenase S245A, A327P mutant complexed with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, L-lactate dehydrogenase, ... | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
2LK0
 
 | |
2L02
 
 | | Solution NMR Structure of protein BT2368 from Bacteroides thetaiotaomicron, Northeast Structural Genomics Consortium Target BtR375 | | Descriptor: | Uncharacterized protein | | Authors: | Eletsky, A, Lee, H, Wang, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-29 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of protein BT2368 from Bacteroides thetaiotaomicron
To be Published
|
|
1TFA
 
 | | OVOTRANSFERRIN, N-TERMINAL LOBE, APO FORM | | Descriptor: | PROTEIN (OVOTRANSFERRIN), SULFATE ION | | Authors: | Mizutani, K, Yamashita, H, Mikami, B, Hirose, M. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternative structural state of transferrin. The crystallographic analysis of iron-loaded but domain-opened ovotransferrin N-lobe.
J.Biol.Chem., 274, 1999
|
|
6RFL
 
 | | Structure of the complete Vaccinia DNA-dependent RNA polymerase complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Grimm, C, Hillen, S.H, Bedenk, K, Bartuli, J, Neyer, S, Zhang, Q, Huettenhofer, A, Erlacher, M, Dienemann, C, Schlosser, A, Urlaub, H, Boettcher, B, Szalay, A.A, Cramer, P, Fischer, U. | | Deposit date: | 2019-04-15 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Vaccinia RNA Polymerase Complexes.
Cell, 179, 2019
|
|
8U9G
 
 | | Human Class I MHC HLA-A2 bound to sorting nexin 24 (127-135) neoantigen KLSHQLVLL | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Arbuiso, A, Weiss, L.I, Brambley, C.A, Ma, J, Keller, G.L.J, Ayres, C.M, Baker, B.M. | | Deposit date: | 2023-09-19 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Accurate modeling of peptide-MHC structures with AlphaFold.
Structure, 32, 2024
|
|
6XBC
 
 | |
6RHC
 
 | | Fragment AZ-003 binding at the TAZpS89/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-azanyl-4-phenyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-19 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
3EF9
 
 | | Replacement of Val3 in Human Thymidylate Synthase Affects Its Kinetic Properties and Intracellular Stability | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Huang, X, Gibson, L.M, Bell, B.J, Lovelace, L.L, Pena, M.M, Berger, F.G, Berger, S.H. | | Deposit date: | 2008-09-08 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Replacement of Val3 in human thymidylate synthase affects its kinetic properties and intracellular stability .
Biochemistry, 49, 2010
|
|
7DNQ
 
 | | Crystal structure of M.tuberculosis imidazole glycerol phosphate dehydratase in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-(1H-1,2,4-triazol-5-ylsulfanyl)ethanamine, CHLORIDE ION, ... | | Authors: | Tiwari, S, Pal, R.K, Biswal, B.K. | | Deposit date: | 2020-12-10 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of M.tuberculosis imidazole glycerol phosphate dehydratase in complex with an inhibitor
To Be Published
|
|
1T76
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with a WxxVW motif | | Descriptor: | 1,2-ETHANEDIOL, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, ... | | Authors: | Hur, E, Pfaff, S.J, Payne, E.S, Gron, H, Buehrer, B.M, Fletterick, R.J. | | Deposit date: | 2004-05-08 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Recognition and accommodation at the androgen receptor coactivator binding interface.
Plos Biol., 2, 2004
|
|
7DP4
 
 | | Crystal structure of wild type Brugia malayi thymidylate synthase complexed with 2'-deoxyuridine monophosphate and methotrexate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, BETA-MERCAPTOETHANOL, METHOTREXATE, ... | | Authors: | Keyunratsami, K, Nualnoi, T, Wongkamchai, S, Songsiriritthigul, C, Chen, C.-J, Canyuk, B. | | Deposit date: | 2020-12-18 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic investigation and inhibition of Brugia malayi thymidylate synthase by a folate analog
To Be Published
|
|
6V33
 
 | | X-ray structure of a sugar N-formyltransferase from Pseudomonas congelans | | Descriptor: | 1,2-ETHANEDIOL, FOLIC ACID, dTDP-4-amino-4,6-dideoxyglucose, ... | | Authors: | Girardi, N.M, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-11-25 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Misannotations of the genes encoding sugar N-formyltransferases.
Protein Sci., 29, 2020
|
|
6RKI
 
 | | Fragment AZ-023 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-[(3-aminophenyl)amino]-4-phenyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
5MD0
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=5, twist=6 | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
1T7R
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with a FxxLF motif | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, FxxLF motif peptide | | Authors: | Hur, E, Pfaff, S.J, Payne, E.S, Gron, H, Buehrer, B.M, Fletterick, R.J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Recognition and accommodation at the androgen receptor coactivator binding interface.
Plos Biol., 2, 2004
|
|
