2CXE
 
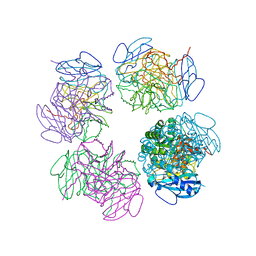 | | Crystal structure of octameric ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Pyrococcus horikoshii OT3 (form-2 crystal) | | Descriptor: | Ribulose bisphosphate carboxylase | | Authors: | Mizohata, E, Mishima, C, Akasaka, R, Uda, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-28 | | Release date: | 2005-12-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of octameric ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Pyrococcus horikoshii OT3 (form-2 crystal)
To be Published
|
|
1QXL
 
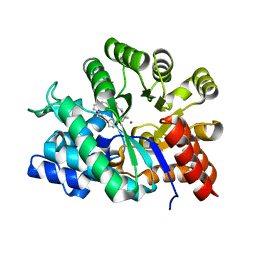 | | Crystal structure of Adenosine deaminase complexed with FR235380 | | Descriptor: | 1-((1R)-1-(HYDROXYMETHYL)-3-{6-[(5-PHENYLPENTANOYL)AMINO]-1H-INDOL-1-YL}PROPYL)-1H-IMIDAZOLE-4-CARBOXAMIDE, Adenosine deaminase, ZINC ION | | Authors: | Kinoshita, T. | | Deposit date: | 2003-09-08 | | Release date: | 2004-09-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based design, synthesis, and structure-activity relationship studies of novel non-nucleoside adenosine deaminase inhibitors
J.Med.Chem., 47, 2004
|
|
2FM4
 
 | |
1QF4
 
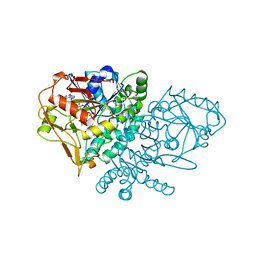 | | DESIGN, SYNTHESIS, AND X-RAY CRYSTAL STRUCTURE OF AN ENZYME BOUND BISUBSTRATE HYBRID INHIBITOR OF ADENYLOSUCCINATE SYNTHETASE | | Descriptor: | (C8-R)-HYDANTOCIDIN 5'-PHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hanessian, S, Lu, P.-P, Sanceau, J.-Y, Chemla, P, Gohda, K, Fonne-Pfister, R, Prade, L, Cowan-Jacob, S.W. | | Deposit date: | 1999-04-06 | | Release date: | 1999-12-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An enzyme-bound bisubstrate hybrid inhibitor of adenylosuccinate synthetase
Angew.Chem.Int.Ed.Engl., 38, 1999
|
|
2D69
 
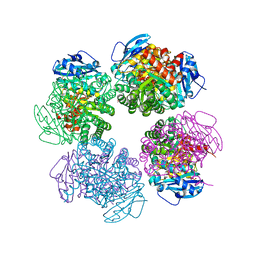 | | Crystal structure of the complex of sulfate ion and octameric ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Pyrococcus horikoshii OT3 (form-2 crystal) | | Descriptor: | Ribulose bisphosphate carboxylase, SULFATE ION | | Authors: | Mizohata, E, Mishima, C, Akasaka, R, Uda, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-11-10 | | Release date: | 2006-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of sulfate ion and octameric ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Pyrococcus horikoshii OT3 (form-2 crystal)
To be Published
|
|
2D37
 
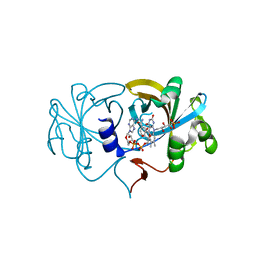 | | The Crystal Structure of Flavin Reductase HpaC complexed with NAD+ | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
2D36
 
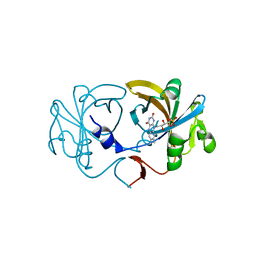 | | The Crystal Structure of Flavin Reductase HpaC | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
2FET
 
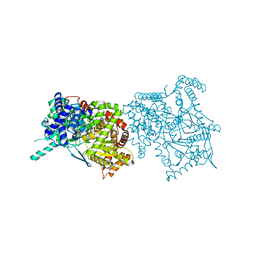 | | Synthesis of C-D-Glycopyranosyl-Hydroquinones and-Benzoquinones. Inhibition of PTP1B. Inhibition of and binding to glycogen phosphorylase in the crystal | | Descriptor: | (1S)-1,5-anhydro-1-(2,5-dihydroxyphenyl)-D-glucitol, Glycogen phosphorylase, muscle form, ... | | Authors: | Chrysina, E.D, Kosmopoulou, M.N, Leonidas, D.D, Oikonomakos, N.G. | | Deposit date: | 2005-12-16 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | In the Search of Glycogen Phosphorylase Inhibitors: Synthesis of C-D-Glycopyranosylbenzo(hydro)quinones Inhibition of and Binding to Glycogen Phosphorylase in the Crystal
Eur.J.Org.Chem., 4, 2007
|
|
2F3Q
 
 | |
2FFR
 
 | | Crystallographic studies on N-azido-beta-D-glucopyranosylamine, an inhibitor of glycogen phosphorylase: comparison with N-acetyl-beta-D-glucopyranosylamine | | Descriptor: | Glycogen phosphorylase, muscle form, N-(azidoacetyl)-beta-D-glucopyranosylamine, ... | | Authors: | Petsalakis, E.I, Chrysina, E.D, Tiraidis, C, Hadjiloi, T, Leonidas, D.D, Oikonomakos, N.G, Aich, U, Varghese, B, Loganathan, D. | | Deposit date: | 2005-12-20 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystallographic studies on N-azidoacetyl-beta-d-glucopyranosylamine, an inhibitor of glycogen phosphorylase: Comparison with N-acetyl-beta-d-glucopyranosylamine.
Bioorg.Med.Chem., 14, 2006
|
|
2DY0
 
 | |
2F3P
 
 | |
1SET
 
 | |
1DGM
 
 | | CRYSTAL STRUCTURE OF ADENOSINE KINASE FROM TOXOPLASMA GONDII | | Descriptor: | ACETIC ACID, ADENOSINE, ADENOSINE KINASE, ... | | Authors: | Cook, W.J, DeLucas, L.J, Chattopadhyay, D. | | Deposit date: | 1999-11-24 | | Release date: | 2000-11-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of adenosine kinase from Toxoplasma gondii at 1.8 A resolution.
Protein Sci., 9, 2000
|
|
1EE1
 
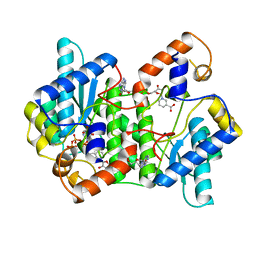 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS COMPLEXED WITH ONE MOLECULE ATP, TWO MOLECULES DEAMIDO-NAD+ AND ONE MG2+ ION | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) SYNTHETASE, ... | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Jedrzejas, M, Brouillette, C, Brouillette, W, Muccio, D, Chattopadhyay, D, Delucas, L. | | Deposit date: | 2000-01-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1EM6
 
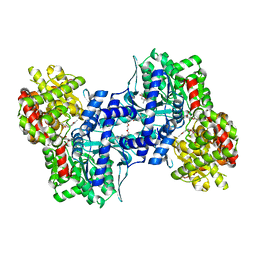 | | HUMAN LIVER GLYCOGEN PHOSPHORYLASE A COMPLEXED WITH GLCNAC AND CP-526,423 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BIS[5-CHLORO-1H-INDOL-2-YL-CARBONYL-AMINOETHYL]-ETHYLENE GLYCOL, LIVER GLYCOGEN PHOSPHORYLASE, ... | | Authors: | Rath, V.L, Ammirati, M, Danley, D.E, Ekstrom, J.L, Hynes, T.R, Olson, T.V, Hoover, D.J. | | Deposit date: | 2000-03-16 | | Release date: | 2000-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human liver glycogen phosphorylase inhibitors bind at a new allosteric site.
Chem.Biol., 7, 2000
|
|
1EXV
 
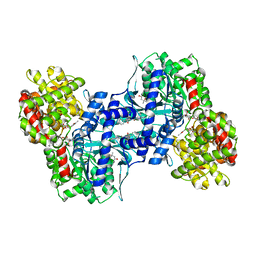 | | HUMAN LIVER GLYCOGEN PHOSPHORYLASE A COMPLEXED WITH GLCNAC AND CP-403,700 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, LIVER GLYCOGEN PHOSPHORYLASE, N-acetyl-beta-D-glucopyranosylamine, ... | | Authors: | Rath, V.L, Ammirati, M, Danley, D.E, Ekstrom, J.L, Hynes, T.R, Olson, T.V, Hoover, D.J. | | Deposit date: | 2000-05-04 | | Release date: | 2000-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human liver glycogen phosphorylase inhibitors bind at a new allosteric site.
Chem.Biol., 7, 2000
|
|
1FS4
 
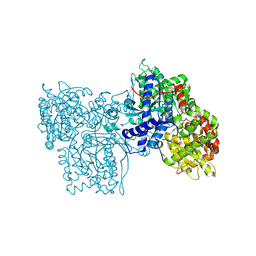 | | Structures of glycogen phosphorylase-inhibitor complexes and the implications for structure-based drug design | | Descriptor: | 1-DEOXY-1-METHOXYCARBAMIDO-BETA-D-GLUCO-2-HEPTULOPYRANOSONAMIDE, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-08 | | Release date: | 2000-10-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
1FBH
 
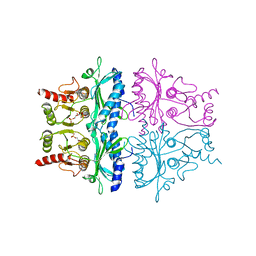 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 1,6-di-O-phosphono-alpha-D-fructofuranose, 1,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
1FBE
 
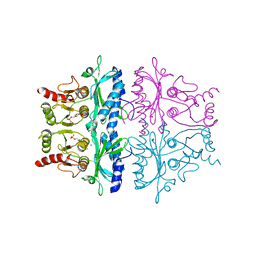 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, FRUCTOSE 1,6-BISPHOSPHATASE, ZINC ION | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
1FBD
 
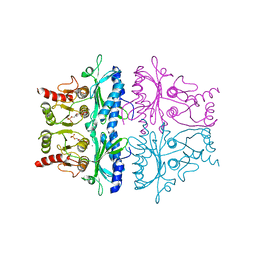 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, FRUCTOSE 1,6-BISPHOSPHATASE, MANGANESE (II) ION | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
1FTW
 
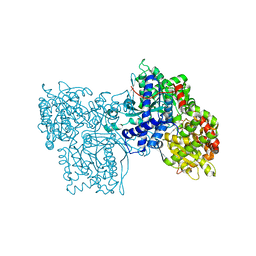 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | Descriptor: | (5S,7R,8S,9S,10R)-3,8,9,10-tetrahydroxy-7-(hydroxymethyl)-6-oxa-1,3-diazaspiro[4.5]decane-2,4-dione, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-13 | | Release date: | 2000-10-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
1GIN
 
 | | CRYSTAL STRUCTURE OF ADENYLOSUCCINATE SYNTHETASE FROM ESCHERICHIA COLI COMPLEXED WITH GDP, IMP, HADACIDIN, NO3-, AND MG2+. DATA COLLECTED AT 298K (PH 6.5). | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE, HADACIDIN, ... | | Authors: | Poland, B.W, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1996-06-18 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of adenylosuccinate synthetase from Escherichia coli complexed with GDP, IMP hadacidin, NO3-, and Mg2+.
J.Mol.Biol., 264, 1996
|
|
1FTQ
 
 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | Descriptor: | (5S,7R,8S,9S,10R)-3-amino-8,9,10-trihydroxy-7-(hydroxymethyl)-6-oxa-1,3-diazaspiro[4.5]decane-2,4-dione, GLYCOGEN PHOSPHORYLASE, INOSINIC ACID, ... | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-13 | | Release date: | 2000-10-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
1FX4
 
 | |
