1NUY
 
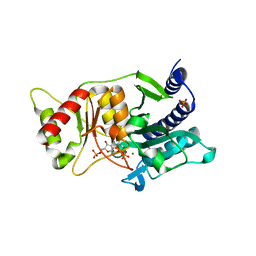 | | Fructose-1,6-Bisphosphatase Complex with Magnesium, Fructose-6-Phosphate, and Phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, MAGNESIUM ION, ... | | Authors: | Choe, J, Iancu, C.V, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2003-02-01 | | Release date: | 2003-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Metaphosphate in the active site of fructose-1,6-bisphosphatase
J.BIOL.CHEM., 278, 2003
|
|
1NUS
 
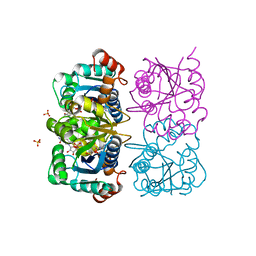 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE COMPLEXED WITH ATP ANALOG AND NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, FKSG76, ... | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
7EZP
 
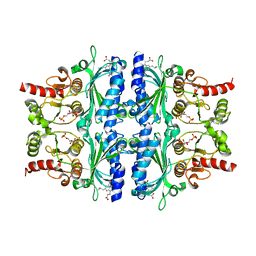 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-(3-hydroxy-3-oxopropyl)-5-(2-methylpropyl)-7-nitro-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZF
 
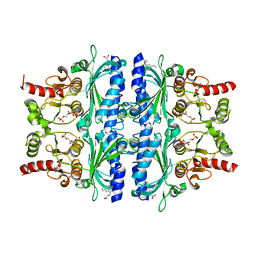 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 7-chloranyl-5-ethyl-3-(3-hydroxy-3-oxopropyl)-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZR
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 5-ethyl-7-nitro-3-[3-oxidanylidene-3-(thiophen-2-ylsulfonylamino)propyl]-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
1P4G
 
 | | Crystal structure of glycogen phosphorylase b in complex with C-(1-azido-alpha-D-glucopyranosyl)formamide | | Descriptor: | C-(1-AZIDO-ALPHA-D-GLUCOPYRANOSYL) FORMAMIDE, Glycogen phosphorylase, muscle form, ... | | Authors: | Chrysina, E.D, Oikonomakos, N.G, Zographos, S.E, Kosmopoulou, M.N, Bischler, N, Leonidas, D.D, Nagy, V, Somsak, L, Gergely, P, Praly, J.-P. | | Deposit date: | 2003-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crsytallographic Studies on alpha- and beta-D-glucopyranosyl Formamide Analogues, Inhibitors of Glycogen Phosphorylase
Biocatal.Biotransfor., 21, 2003
|
|
1P2B
 
 | | Crystal Structure of Glycogen Phosphorylase B in Complex with Maltoheptaose | | Descriptor: | Glycogen phosphorylase, muscle form, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Pinotsis, N, Leonidas, D.D, Chrysina, E.D, Oikonomakos, N.G, Mavridis, I.M. | | Deposit date: | 2003-04-15 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The binding of beta- and gamma-cyclodextrins to glycogen phosphorylase b: Kinetic and crystallographic studies.
Protein Sci., 12, 2003
|
|
2CWX
 
 | | Crystal structure of octameric ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Pyrococcus horikoshii OT3 (form-1 crystal) | | Descriptor: | Ribulose bisphosphate carboxylase | | Authors: | Mizohata, E, Mishima, C, Akasaka, R, Uda, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-27 | | Release date: | 2005-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of octameric ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Pyrococcus horikoshii OT3 (form-1 crystal)
To be Published
|
|
7D5C
 
 | | IleRS in complex with a tRNA site inhibitor | | Descriptor: | (2E,4S,5S,6E,8E)-10-[(2S,3R,6S,8R,9S)-3-butyl-9-methyl-2-[(1E,3E)-3-methyl-5-oxidanyl-5-oxidanylidene-penta-1,3-dienyl]-3-(4-oxidanyl-4-oxidanylidene-butanoyl)oxy-1,7-dioxaspiro[5.5]undecan-8-yl]-4,8-dimethyl-5-oxidanyl-deca-2,6,8-trienoic acid, 1,2-ETHANEDIOL, Isoleucine--tRNA ligase, ... | | Authors: | Chen, B, Luo, S, Zhou, H. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Inhibitory mechanism of reveromycin A at the tRNA binding site of a class I synthetase.
Nat Commun, 12, 2021
|
|
1NUP
 
 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE COMPLEX WITH NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, FKSG76, SULFATE ION | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
1NYR
 
 | | Structure of Staphylococcus aureus threonyl-tRNA synthetase complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, THREONINE, ZINC ION, ... | | Authors: | Torres-Larios, A, Sankaranarayanan, R, Rees, B, Dock-Bregeon, A.C, Moras, D. | | Deposit date: | 2003-02-13 | | Release date: | 2003-10-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational movements and cooperativity upon amino acid, ATP and tRNA binding in threonyl-tRNA synthetase
J.Mol.Biol., 331, 2003
|
|
1PL0
 
 | | Crystal structure of human ATIC in complex with folate-based inhibitor, BW2315U89UC | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Bifunctional purine biosynthesis protein PURH, N-(4-{[(2-AMINO-4-OXO-3,4-DIHYDROQUINAZOLIN-6-YL)AMINO]SULFONYL}BENZOYL)GLUTAMIC ACID, ... | | Authors: | Cheong, C.G, Greasley, S.E, Horton, P.A, Beardsley, G.P, Wilson, I.A. | | Deposit date: | 2003-06-06 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Human Bifunctional Enzyme Aminoimidazole-4-carboxamide Ribonucleotide Transformylase/IMP Cyclohydrolase in Complex with Potent Sulfonyl-containing Antifolates.
J.Biol.Chem., 279, 2004
|
|
2E28
 
 | |
5OEZ
 
 | | Crystal structure of Leishmania major fructose-1,6-bisphosphatase in apo form. | | Descriptor: | FBP protein | | Authors: | Yuan, M, Vasquez-Valdivieso, M.G, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2017-07-10 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structures of Leishmania Fructose-1,6-Bisphosphatase Reveal Species-Specific Differences in the Mechanism of Allosteric Inhibition.
J. Mol. Biol., 429, 2017
|
|
2E1W
 
 | | Crystal structure of adenosine deaminase complexed with potent inhibitors | | Descriptor: | 1-{(1R,2S)-2-HYDROXY-1-[2-(1-NAPHTHYL)ETHYL]PROPYL}-1H-IMIDAZOLE-4-CARBOXAMIDE, Adenosine deaminase, ZINC ION | | Authors: | Kinoshita, T. | | Deposit date: | 2006-10-30 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design and Synthesis of Non-Nucleoside, Potent, and Orally Bioavailable Adenosine Deaminase Inhibitors
J.Med.Chem., 47, 2004
|
|
2AZW
 
 | | Crystal structure of the MutT/nudix family protein from Enterococcus faecalis | | Descriptor: | MutT/nudix family protein, PENTAETHYLENE GLYCOL | | Authors: | Zhang, R, Zhou, M, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-12 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9A crystal structure of the MutT/nudix family protein from Enterococcus faecalis
To be Published
|
|
1OBH
 
 | | LEUCYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS COMPLEXED WITH A PRE-TRANSFER EDITING SUBSTRATE ANALOGUE IN BOTH SYNTHETIC ACTIVE SITE AND EDITING SITE | | Descriptor: | LEUCYL-TRNA SYNTHETASE, MERCURY (II) ION, NORVALINE, ... | | Authors: | Cusack, S, Yaremchuk, A, Tukalo, M. | | Deposit date: | 2003-01-31 | | Release date: | 2003-05-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Basis of Pre- and Posttransfer Editing by Leucyl-tRNA Synthetase
Mol.Cell, 11, 2003
|
|
2AMV
 
 | | THE STRUCTURE OF GLYCOGEN PHOSPHORYLASE B WITH AN ALKYL-DIHYDROPYRIDINE-DICARBOXYLIC ACID | | Descriptor: | 2,3-DICARBOXY-4-(2-CHLORO-PHENYL)-1-ETHYL-5-ISOPROPOXYCARBONYL-6-METHYL-PYRIDINIUM, GLYCEROL, PROTEIN (GLYCOGEN PHOSPHORYLASE), ... | | Authors: | Zographos, S.E, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of glycogen phosphorylase b with an alkyldihydropyridine-dicarboxylic acid compound, a novel and potent inhibitor.
Structure, 5, 1997
|
|
1O5R
 
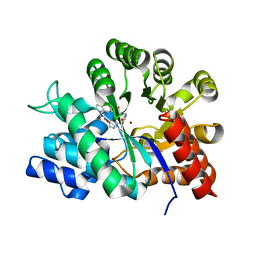 | | Crystal structure of adenosine deaminase complexed with a potent inhibitor | | Descriptor: | 1-[(1R)-3-(6-{[(BENZYLAMINO)CARBONYL]AMINO}-1H-INDOL-1-YL)-1-(HYDROXYMETHYL)PROPYL]-1H-IMIDAZOLE-4-CARBOXAMIDE, Adenosine deaminase, ZINC ION | | Authors: | Kinoshita, T. | | Deposit date: | 2003-10-05 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-based design, synthesis, and structure-activity relationship studies of novel non-nucleoside adenosine deaminase inhibitors
J.Med.Chem., 47, 2004
|
|
1OBC
 
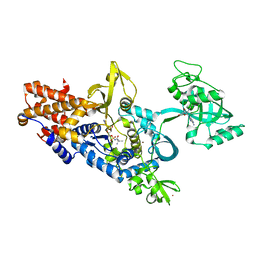 | | LEUCYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS COMPLEXED WITH A POST-TRANSFER EDITING SUBSTRATE ANALOGUE | | Descriptor: | 2'-AMINO-2'-DEOXYADENOSINE, LEUCINE, LEUCYL-TRNA SYNTHETASE, ... | | Authors: | Cusack, S, Yaremchuk, A, Tukalo, M. | | Deposit date: | 2003-01-30 | | Release date: | 2003-05-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Basis of Pre- and Posttransfer Editing by Leucyl-tRNA Synthetase
Mol.Cell, 11, 2003
|
|
7CVN
 
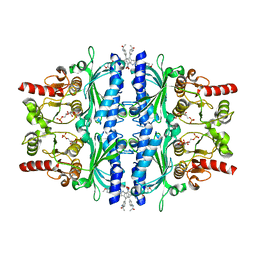 | | The N-arylsulfonyl-indole-2-carboxamide-based inhibitors against fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4-(3-acetamidophenyl)-N-(4-methoxyphenyl)sulfonyl-7-nitro-1H-indole-2-carboxamide, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X, Zhou, J, Xu, B. | | Deposit date: | 2020-08-26 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Design,synthesis,biological evaluation and binding mode analysis of 7-nitro-indole-N-acylarylsulfonamide-based fructose-1,6-bisphosphatase inhibitors
Chinese journal of medicinal chemistry, 30, 2020
|
|
1QFJ
 
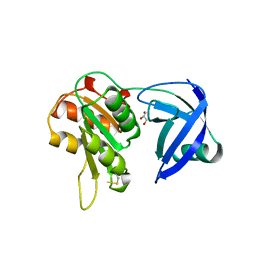 | | CRYSTAL STRUCTURE OF NAD(P)H:FLAVIN OXIDOREDUCTASE FROM ESCHERICHIA COLI | | Descriptor: | GLYCEROL, PROTEIN (FLAVIN REDUCTASE) | | Authors: | Ingelman, M, Ramaswamy, S, Niviere, V, Fontecave, M, Eklund, H. | | Deposit date: | 1999-04-12 | | Release date: | 1999-06-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of NAD(P)H:flavin oxidoreductase from Escherichia coli.
Biochemistry, 38, 1999
|
|
7DBS
 
 | |
1Q33
 
 | | Crystal structure of human ADP-ribose pyrophosphatase NUDT9 | | Descriptor: | ADP-ribose pyrophosphatase, SULFATE ION, beta-D-glucopyranose | | Authors: | Shen, B.W, Perraud, A.L, Scharenberg, A, Stoddard, B.L. | | Deposit date: | 2003-07-28 | | Release date: | 2003-09-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Crystal Structure and Mutational Analysis of Human NUDT9
J.Mol.Biol., 332, 2003
|
|
1NUR
 
 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE | | Descriptor: | FKSG76, SULFATE ION | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
