7G8Y
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with Z1449748885 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, N-(4-methoxyphenyl)glycinamide, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
3BD8
 
 | | Glucogen Phosphorylase complex with 1(-D-glucopyranosyl) cytosine | | Descriptor: | 4-amino-1-beta-D-glucopyranosylpyrimidin-2(1H)-one, Glycogen phosphorylase, muscle form, ... | | Authors: | Sovantzis, D.A, Hadjiloi, T, Hayes, J.M, Zographos, S.E, Chrysina, E.D, Oikonomakos, N.G. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | D-Glucopyranosyl pyrimidine nucleoside binding to muscle glycogen phosphorylase b
To be Published
|
|
3BCR
 
 | | Glycogen Phosphorylase b in complex with AZT | | Descriptor: | 3'-azido-3'-deoxythymidine, Glycogen phosphorylase, muscle form | | Authors: | Sovantzis, D.A, Hadjiloi, T, Hayes, J.M, Zographos, S.E, Chrysina, E.D, Oikonomakos, N.G. | | Deposit date: | 2007-11-13 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | D-Glucopyranosyl pyrimidine nucleoside binding to muscle glycogen phosphorylase b
To be Published
|
|
3BD7
 
 | | Glycogen Phosphorylase complex with 1(-D-glucopyranosyl) thymine | | Descriptor: | 1-beta-D-glucopyranosyl-5-methylpyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form, ... | | Authors: | Sovantzis, D.A, Hadjiloi, T, Hayes, J.M, Zographos, S.E, Chrysina, E.D, Oikonomakos, N.G. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D-Glucopyranosyl pyrimidine nucleoside binding to muscle glycogen phosphorylase b
To be Published
|
|
3B4W
 
 | | Crystal structure of Mycobacterium tuberculosis aldehyde dehydrogenase complexed with NAD+ | | Descriptor: | Aldehyde dehydrogenase, ETHANOL, GLYCEROL, ... | | Authors: | Moon, J.H, Lyon, A.E, Yu, M, Hung, L.-W, Terwilliger, T, Kim, C.-Y, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-10-24 | | Release date: | 2007-11-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of aldehyde dehydrogenase from Mycobacterium tuberculosis complexed with NAD+.
To Be Published
|
|
3AQN
 
 | |
3AQL
 
 | |
3AQM
 
 | | Structure of bacterial protein (form II) | | Descriptor: | MAGNESIUM ION, Poly(A) polymerase | | Authors: | Toh, Y, Takeshita, D, Tomita , K. | | Deposit date: | 2010-11-09 | | Release date: | 2011-02-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Mechanism for the alteration of the substrate specificities of template-independent RNA polymerases
Structure, 19, 2011
|
|
3BQV
 
 | |
3BQ9
 
 | | Crystal structure of predicted nucleotide-binding protein from Idiomarina baltica OS145 | | Descriptor: | GLYCEROL, Predicted Rossmann fold nucleotide-binding domain-containing protein, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Meyer, A.J, Dickey, M, Eberle, M, Koss, J, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-19 | | Release date: | 2008-01-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Predicted Nucleotide-Binding Protein from Idiomarina baltica.
To be Published
|
|
2ZB2
 
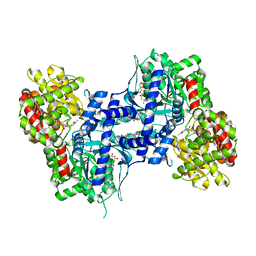 | | Human liver glycogen phosphorylase a complexed with glcose and 5-chloro-N-[4-(1,2-dihydroxyethyl)phenyl]-1H-indole-2-carboxamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-chloro-N-{4-[(1R)-1,2-dihydroxyethyl]phenyl}-1H-indole-2-carboxamide, ... | | Authors: | Katayama, N, Onda, K. | | Deposit date: | 2007-10-15 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthesis of 5-chloro-N-aryl-1H-indole-2-carboxamide derivatives as inhibitors of human liver glycogen phosphorylase a.
Bioorg.Med.Chem., 16, 2008
|
|
7SSB
 
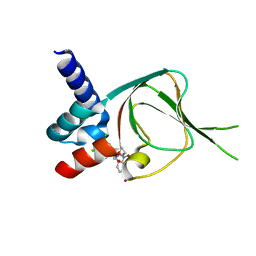 | | Co-structure of PKG1 regulatory domain with compound 33 | | Descriptor: | 4-({(2S,3S)-3-[(1S)-1-(3,5-dichlorophenyl)-2-hydroxyethoxy]-2-phenylpiperidin-1-yl}methyl)-3-nitrobenzoic acid, cGMP-dependent protein kinase 1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2021-11-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Optimization and Mechanistic Investigations of Novel Allosteric Activators of PKG1 alpha.
J.Med.Chem., 65, 2022
|
|
5HD1
 
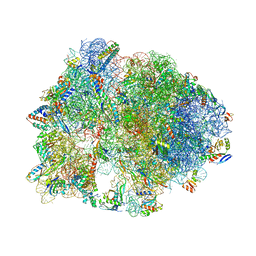 | | Crystal structure of antimicrobial peptide Pyrrhocoricin bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gagnon, M.G, Roy, R.N, Lomakin, I.B, Florin, T, Mankin, A.S, Steitz, T.A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition.
Nucleic Acids Res., 44, 2016
|
|
5HCR
 
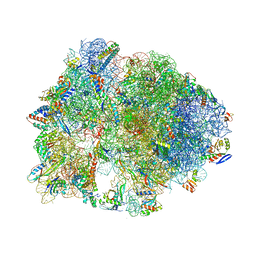 | | Crystal structure of antimicrobial peptide Oncocin 10wt bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gagnon, M.G, Roy, R.N, Lomakin, I.B, Florin, T, Mankin, A.S, Steitz, T.A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition.
Nucleic Acids Res., 44, 2016
|
|
5HCQ
 
 | | Crystal structure of antimicrobial peptide Oncocin d15-19 bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gagnon, M.G, Roy, R.N, Lomakin, I.B, Florin, T, Mankin, A.S, Steitz, T.A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition.
Nucleic Acids Res., 44, 2016
|
|
5HAU
 
 | | Crystal structure of antimicrobial peptide Bac7(1-19) bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gagnon, M.G, Roy, R.N, Lomakin, I.B, Florin, T, Mankin, A.S, Steitz, T.A. | | Deposit date: | 2015-12-30 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition.
Nucleic Acids Res., 44, 2016
|
|
5HCP
 
 | | Crystal structure of antimicrobial peptide Metalnikowin bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gagnon, M.G, Roy, R.N, Lomakin, I.B, Florin, T, Mankin, A.S, Steitz, T.A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition.
Nucleic Acids Res., 44, 2016
|
|
8OKU
 
 | | Salt-Inducible Kinase 3 in complex with an inhibitor | | Descriptor: | Serine/threonine-protein kinase SIK3, ~{N}-ethyl-4-[5-[1-(2-hydroxyethyl)pyrazol-4-yl]benzimidazol-1-yl]-2,6-dimethoxy-benzamide | | Authors: | Flower, T.G, Leonard, P.M, Lamers, M.B.A.C, Mollat, P. | | Deposit date: | 2023-03-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Optimization of Selectivity and Pharmacokinetic Properties of Salt-Inducible Kinase Inhibitors that Led to the Discovery of Pan-SIK Inhibitor GLPG3312.
J.Med.Chem., 67, 2024
|
|
3U0L
 
 | | Crystal structure of the engineered fluorescent protein mRuby, crystal form 1, pH 4.5 | | Descriptor: | ACETATE ION, mRuby | | Authors: | Akerboom, J, Looger, L.L, Schreiter, E.R. | | Deposit date: | 2011-09-28 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics.
Front Mol Neurosci, 6, 2013
|
|
3U0M
 
 | |
8FNY
 
 | | Nucleotide-bound structure of a functional construct of eukaryotic elongation factor 2 kinase. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Piserchio, A, Isiorho, E.A, Dalby, K.N, Ghose, R. | | Deposit date: | 2022-12-28 | | Release date: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | ADP enhances the allosteric activation of eukaryotic elongation factor 2 kinase by calmodulin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FO6
 
 | | Nucleotide-free structure of a functional construct of eukaryotic elongation factor 2 kinase. | | Descriptor: | CALCIUM ION, Calmodulin-1, Eukaryotic elongation factor 2 kinase, ... | | Authors: | Piserchio, A, Isiorho, E.A, Dalby, K.N, Ghose, R. | | Deposit date: | 2022-12-29 | | Release date: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | ADP enhances the allosteric activation of eukaryotic elongation factor 2 kinase by calmodulin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4USV
 
 | |
6K4L
 
 | |
6K4K
 
 | |
