4V4D
 
 | | Crystal Structure of Pyrogallol-Phloroglucinol Transhydroxylase from Pelobacter acidigallici complexed with pyrogallol | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, BENZENE-1,2,3-TRIOL, CALCIUM ION, ... | | Authors: | Messerschmidt, A, Niessen, H, Abt, D, Einsle, O, Schink, B, Kroneck, P.M.H. | | Deposit date: | 2004-06-02 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
4UTM
 
 | | XenA - Reduced - Y183F variant in complex with 8-hydroxycoumarin | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 8-HYDROXYCOUMARIN, SULFATE ION, ... | | Authors: | Werther, T, Dobbek, H. | | Deposit date: | 2014-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Redox-dependent substrate-cofactor interactions in the Michaelis-complex of a flavin-dependent oxidoreductase
Nat Commun, 8, 2017
|
|
4UWD
 
 | |
4UTK
 
 | | XenA - reduced - Y183F variant | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, SULFATE ION, XENOBIOTIC REDUCTASE | | Authors: | Werther, T, Dobbek, H. | | Deposit date: | 2014-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Redox-dependent substrate-cofactor interactions in the Michaelis-complex of a flavin-dependent oxidoreductase
Nat Commun, 8, 2017
|
|
8VLM
 
 | | Crystal structure of the yeast cytosine deaminase (yCD) E64V-M100W heterodimer | | Descriptor: | 1,2-ETHANEDIOL, Cytosine deaminase, ZINC ION | | Authors: | Picard, M.-E, Grenier, G, Despres, P.C, Dube, A.K, Landry, C.R, Shi, R. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Compensatory mutations potentiate constructive neutral evolution by gene duplication.
Science, 385, 2024
|
|
8W06
 
 | | Crystal Structure of the reconstruction of the ancestral triosephosphate isomerase of the last opisthokont common ancestor obtained by maximum likelihood with PGH | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ACETIC ACID, ... | | Authors: | Perez-Nino, J.A, Rodriguez-Romero, A, Guerra-Borrego, Y, Fernandez-Velasco, D.A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Stable monomers in the ancestral sequence reconstruction of the last opisthokont common ancestor of dimeric triosephosphate isomerase.
Protein Sci., 33, 2024
|
|
4WDX
 
 | | 17beta-HSD5 in complex with [4-(2-hydroxyethyl)piperidin-1-yl](5-methyl-1H-indol-2-yl)methanone | | Descriptor: | Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [4-(2-hydroxyethyl)piperidin-1-yl](5-methyl-1H-indol-2-yl)methanone | | Authors: | Amano, Y, Yamaguchi, T. | | Deposit date: | 2014-09-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structures of complexes of type 5 17 beta-hydroxysteroid dehydrogenase with structurally diverse inhibitors: insights into the conformational changes upon inhibitor binding.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2BL8
 
 | | 1.6 Angstrom crystal structure of EntA-im: a bacterial immunity protein conferring immunity to the antimicrobial activity of the pediocin-like bacteriocin, enterocin A | | Descriptor: | CITRATE ANION, ENTEROCINE A IMMUNITY PROTEIN | | Authors: | Johnsen, L, Dalhus, B, Leiros, I, Nissen-Meyer, J. | | Deposit date: | 2005-03-02 | | Release date: | 2005-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6-A Crystal Structure of Enta-Im: A Bacterial Immunity Protein Conferring Immunity to the Antimicrobial Activity of the Pediocin-Like Bacteriocin Enterocin A
J.Biol.Chem., 280, 2005
|
|
2C3D
 
 | | 2.15 Angstrom crystal structure of 2-ketopropyl coenzyme M oxidoreductase carboxylase with a coenzyme M disulfide bound at the active site | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-OXOPROPYL-COM REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pandey, A.S, Nocek, B, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2005-10-05 | | Release date: | 2005-11-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanistic Implications of the Structure of the Mixed-Disulfide Intermediate of the Disulfide Oxidoreductase, 2-Ketopropyl-Coenzyme M Oxidoreductase/Carboxylase.
Biochemistry, 45, 2006
|
|
8VHU
 
 | | Crystal structure of dATP bound E. coli class Ia ribonucleotide reductase alpha construct fused with the C-terminal tail of E. coli class Ia beta subunit | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CHLORIDE ION, Fusion protein of Ribonucleoside-diphosphate reductase 1 subunits alpha and beta, ... | | Authors: | Funk, M.A, Zimanyi, C.M, Drennan, C.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How ATP and dATP Act as Molecular Switches to Regulate Enzymatic Activity in the Prototypical Bacterial Class Ia Ribonucleotide Reductase.
Biochemistry, 63, 2024
|
|
8VHO
 
 | | Crystal Structure of E. coli class Ia ribonucleotide reductase alpha subunit bound to dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Funk, M.A, Zimanyi, C.M, Drennan, C.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | How ATP and dATP Act as Molecular Switches to Regulate Enzymatic Activity in the Prototypical Bacterial Class Ia Ribonucleotide Reductase.
Biochemistry, 63, 2024
|
|
8VHN
 
 | | Crystal Structure of E. coli class Ia ribonucleotide reductase alpha subunit bound to two ATP molecules | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Funk, M.A, Zimanyi, C.M, Drennan, C.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | How ATP and dATP Act as Molecular Switches to Regulate Enzymatic Activity in the Prototypical Bacterial Class Ia Ribonucleotide Reductase.
Biochemistry, 63, 2024
|
|
8VU3
 
 | | Cryo-EM structure of cyanobacterial PSI with bound platinum nanoparticles | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Gisriel, C.J, Malavath, T, Brudvig, G.W, Utschig, L.M. | | Deposit date: | 2024-01-28 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Structure of a biohybrid photosystem I-platinum nanoparticle solar fuel catalyst.
Nat Commun, 15, 2024
|
|
2C3C
 
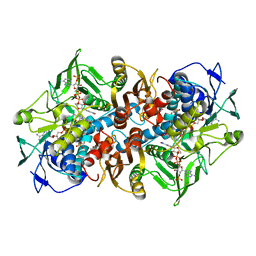 | | 2.01 Angstrom X-ray crystal structure of a mixed disulfide between coenzyme M and NADPH-dependent oxidoreductase 2-ketopropyl coenzyme M carboxylase | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-OXOPROPYL-COM REDUCTASE, ACETONE, ... | | Authors: | Pandey, A.S, Nocek, B, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2005-10-05 | | Release date: | 2005-12-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanistic Implications of the Structure of the Mixed-Disulfide Intermediate of the Disulfide Oxidoreductase, 2-Ketopropyl-Coenzyme M Oxidoreductase/Carboxylase.
Biochemistry, 45, 2006
|
|
8VQ2
 
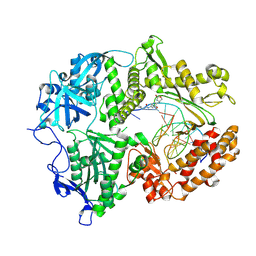 | | HSV1 polymerase ternary complex with dsDNA and compound 44 | | Descriptor: | 2-(4-bromophenyl)-N-(3-methoxy-4-{[(4S)-2-oxo-1,3-oxazolidin-4-yl]methyl}phenyl)acetamide, DNA (5'-D(P*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*TP*AP*C)-3'), DNA (5'-D(P*TP*GP*GP*TP*AP*GP*GP*GP*GP*AP*AP*GP*GP*AP*T)-3'), ... | | Authors: | Hayes, R.P, Heo, M.R, Plotkin, M. | | Deposit date: | 2024-01-17 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (3.829 Å) | | Cite: | Discovery of Broad-Spectrum Herpes Antiviral Oxazolidinone Amide Derivatives and Their Structure-Activity Relationships.
Acs Med.Chem.Lett., 15, 2024
|
|
5VDC
 
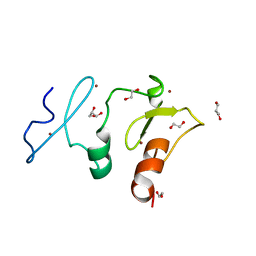 | | Crystal structure of the human DPF2 tandem PHD finger domain | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, ZINC ION, ... | | Authors: | Huber, F.M, Davenport, A.M, Hoelz, A. | | Deposit date: | 2017-04-01 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Histone-binding of DPF2 mediates its repressive role in myeloid differentiation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VFB
 
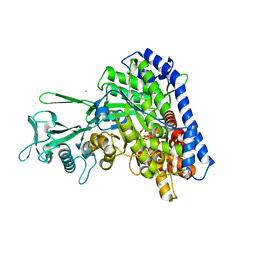 | | 1.36 Angstrom Resolution Crystal Structure of Malate Synthase G from Pseudomonas aeruginosa in Complex with Glycolic Acid. | | Descriptor: | CHLORIDE ION, GLYCOLIC ACID, Malate synthase G, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-07 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | 1.36 Angstrom Resolution Crystal Structure of Malate Synthase G from Pseudomonas aeruginosa in Complex with Glycolic Acid.
To Be Published
|
|
5IO3
 
 | | Crystal structure of the legionella pneumophila effector protein RavZ - I422 | | Descriptor: | Uncharacterized protein RavZ | | Authors: | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | Deposit date: | 2016-03-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
1KS9
 
 | | Ketopantoate Reductase from Escherichia coli | | Descriptor: | 2-DEHYDROPANTOATE 2-REDUCTASE | | Authors: | Matak-Vinkovic, D, Vinkovic, M, Saldanha, S.A, Ashurst, J.A, von Delft, F, Inoue, T, Miguel, R.N, Smith, A.G, Blundell, T.L, Abell, C. | | Deposit date: | 2002-01-11 | | Release date: | 2002-01-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Escherichia coli ketopantoate reductase at 1.7 A resolution and insight into the enzyme mechanism.
Biochemistry, 40, 2001
|
|
5I9U
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
9CXO
 
 | | Crystal structure of Human FN3K(D217S) mutant bound with ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Garg, A, On, K.F, Joshua-tor, L. | | Deposit date: | 2024-07-31 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The molecular basis of Human FN3K mediated phosphorylation of glycated substrates.
Nat Commun, 16, 2025
|
|
1K5P
 
 | | Hydrolytic haloalkane dehalogenase LINB from sphingomonas paucimobilis UT26 at 1.8A resolution | | Descriptor: | 1,3,4,6-tetrachloro-1,4-cyclohexadiene hydrolase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Streltsov, V.A, Damborsky, J, Wilce, M.C.J. | | Deposit date: | 2001-10-12 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates
Biochemistry, 42, 2003
|
|
5VBC
 
 | | Crystal structure of ATXR5 in complex with histone H3.1 | | Descriptor: | DIMETHYL SULFOXIDE, Histone H3.1 peptide, Probable Histone-lysine N-methyltransferase ATXR5, ... | | Authors: | Couture, J.-F, Bergamin, E. | | Deposit date: | 2017-03-29 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
4DJN
 
 | |
3SZL
 
 | | IspH:Ligand Mutants - wt 70sec | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Span, I, Graewert, T, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Mutant IspH Proteins Reveal a Rotation of the Substrate's Hydroxymethyl Group during Catalysis.
J.Mol.Biol., 416, 2012
|
|
