8B5Q
 
 | |
8BV9
 
 | | Acylphosphatase from E. coli | | Descriptor: | 1,2-ETHANEDIOL, Acylphosphatase, GLYCEROL, ... | | Authors: | Gavira, J.A, Martinez-Rodriguez, S. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | First 3-D structural evidence of a native-like intertwined dimer in the acylphosphatase family.
Biochem.Biophys.Res.Commun., 682, 2023
|
|
8B62
 
 | | Crystal Structure of P. aeruginosa WaaG in complex with UDP-galactose | | Descriptor: | GALACTOSE-URIDINE-5'-DIPHOSPHATE, GLYCEROL, UDP-glucose:(Heptosyl) LPS alpha 1,3-glucosyltransferase WaaG | | Authors: | Scaletti, E, Gustafsson Westergren, R, Stenmark, P. | | Deposit date: | 2022-09-25 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
J.Biol.Chem., 299, 2023
|
|
8B63
 
 | | Crystal Structure of P. aeruginosa WaaG in complex with UDP-GalNAc | | Descriptor: | ACETATE ION, UDP-glucose:(Heptosyl) LPS alpha 1,3-glucosyltransferase WaaG, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Scaletti, E, Gustafsson Westergren, R, Stenmark, P. | | Deposit date: | 2022-09-25 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
J.Biol.Chem., 299, 2023
|
|
8BFG
 
 | | Solution structure of human apo/Calmodulin G113R (G114R) | | Descriptor: | Calmodulin-1 | | Authors: | Wimmer, R, Holler, C.V, Petersson, N.M, Brohus, M.B, Niemelae, M, Overgaard, M.T, Iwai, H. | | Deposit date: | 2022-10-25 | | Release date: | 2023-10-04 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Allosteric changes in protein stability and dynamics as pathogenic mechanism for calmodulin variants not affecting Ca 2+ coordinating residues.
Cell Calcium, 117, 2023
|
|
4GU3
 
 | |
3N1V
 
 | | Human FPPS COMPLEX WITH FBS_01 | | Descriptor: | (5-chloro-3-methyl-1-benzothiophen-2-yl)acetic acid, FARNESYL PYROPHOSPHATE SYNTHASE, PHOSPHATE ION | | Authors: | Rondeau, J.-M. | | Deposit date: | 2010-05-17 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Allosteric non-bisphosphonate FPPS inhibitors identified by fragment-based discovery.
Nat.Chem.Biol., 6, 2010
|
|
8B9X
 
 | | Chimeric protein of human UFM1 E3 ligase, UFL1, and DDRGK1 | | Descriptor: | CHLORIDE ION, DDRGK domain-containing protein 1,E3 UFM1-protein ligase 1 | | Authors: | Wiener, R, Isupov, M, banerjee, S. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.066 Å) | | Cite: | Structural study of UFL1-UFC1 interaction uncovers the role of UFL1 N-terminal helix in ufmylation.
Embo Rep., 24, 2023
|
|
3N49
 
 | | Human FPPS COMPLEX WITH NOV_292 | | Descriptor: | FARNESYL PYROPHOSPHATE SYNTHASE, PHOSPHATE ION, naphtho[2,1-b]thiophen-1-ylacetic acid | | Authors: | Rondeau, J.-M. | | Deposit date: | 2010-05-21 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric non-bisphosphonate FPPS inhibitors identified by fragment-based discovery.
Nat.Chem.Biol., 6, 2010
|
|
3NB9
 
 | | Crystal structure of radical H88Q Synechocystis sp. PCYA | | Descriptor: | 1,2-ETHANEDIOL, BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Gae, D.D. | | Deposit date: | 2010-06-03 | | Release date: | 2010-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for hydration dynamics in radical stabilization of bilin reductase mutants.
Biochemistry, 49, 2010
|
|
3N3L
 
 | | Human FPPS complex with FBS_03 | | Descriptor: | (6-methoxy-1-benzofuran-3-yl)acetic acid, FARNESYL PYROPHOSPHATE SYNTHASE, PHOSPHATE ION | | Authors: | Rondeau, J.-M. | | Deposit date: | 2010-05-20 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Allosteric non-bisphosphonate FPPS inhibitors identified by fragment-based discovery.
Nat.Chem.Biol., 6, 2010
|
|
4H6H
 
 | |
3NBZ
 
 | | Crystal structure of the HIV-1 Rev NES-CRM1-RanGTP nuclear export complex (crystal I) | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7ZQ6
 
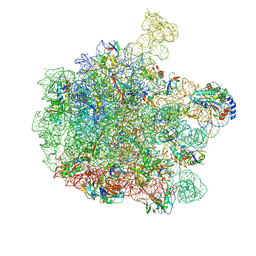 | | 70S E. coli ribosome with truncated uL23 and uL24 loops and a stalled filamin domain 5 nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Wlodarski, T, Ahn, M, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
7ZP8
 
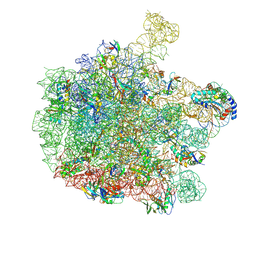 | | 70S E. coli ribosome with a stalled filamin domain 5 nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Plessa, E, Wlodarski, T, Ahn, M, Chan, S.H.S, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
6WW3
 
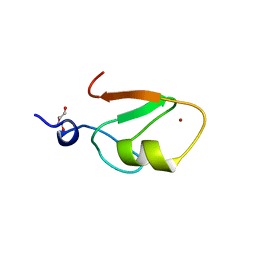 | | Crystal structure of HERC2 ZZ domain in complex with SUMO1 tail | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SUMO1 linked HERC2 ZZ domain (Small ubiquitin-related modifier 1,E3 ubiquitin-protein ligase HERC2), ... | | Authors: | Liu, J, Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2020-05-07 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural Insight into Binding of the ZZ Domain of HERC2 to Histone H3 and SUMO1.
Structure, 28, 2020
|
|
7ZOD
 
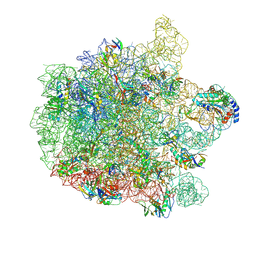 | | 70S E. coli ribosome with an extended uL23 loop from Candidatus marinimicrobia | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Plessa, E, Wlodarski, T, Ahn, M, Sidhu, H, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-04-25 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
3OG5
 
 | | Crystal Structure of BamA POTRA45 tandem | | Descriptor: | Outer membrane protein assembly complex, YaeT protein | | Authors: | Gatzeva-Topalova, P.Z, Warner, L.R, Pardi, A, Sousa, M.C. | | Deposit date: | 2010-08-16 | | Release date: | 2010-11-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure and Flexibility of the Complete Periplasmic Domain of BamA: The Protein Insertion Machine of the Outer Membrane
Structure, 18, 2010
|
|
7ZQ5
 
 | | 70S E. coli ribosome with truncated uL23 and uL24 loops | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Wlodarski, T, Ahn, M, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
8SW5
 
 | |
8SW6
 
 | |
4I0C
 
 | | The structure of the camelid antibody cAbHuL5 in complex with human lysozyme | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | De Genst, E, Chan, P.H, Pardon, E, Kumita, J.R, Christodoulou, J, Menzer, L, Chirgadze, D.Y, Robinson, C.V, Muyldermans, S, Matagne, A, Wyns, L, Dobson, C.M, Dumoulin, M. | | Deposit date: | 2012-11-16 | | Release date: | 2013-10-09 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A nanobody binding to non-amyloidogenic regions of the protein human lysozyme enhances partial unfolding but inhibits amyloid fibril formation.
J.Phys.Chem.B, 117, 2013
|
|
6WX5
 
 | | Adducts formed after 3 weeks in the reaction of chlorido[chlorido(2,2'-((2-([2,2':6',2''-Terpyridin]-4'-yloxy)ethyl)azanediyl)bis(ethan-1-ol))platinum(II)] with HEWL | | Descriptor: | ACETATE ION, Lysozyme, PLATINUM (II) ION, ... | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2020-05-09 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mustards-Derived Terpyridine-Platinum Complexes as Anticancer Agents: DNA Alkylation vs Coordination.
Inorg.Chem., 60, 2021
|
|
4I9U
 
 | | Crystal structure of rabbit LDHA in complex with a fragment inhibitor AP26256 | | Descriptor: | 6-({2-[(5-chloro-2-methoxyphenyl)amino]-2-oxoethyl}sulfanyl)pyridine-3-carboxylic acid, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Kohlmann, A, Stephan, Z.G, Commodore, L, Greenfield, M.T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
8BD2
 
 | | Calcium-bound Calmodulin variant G113R | | Descriptor: | CALCIUM ION, Calmodulin-3 | | Authors: | Wimmer, R, Holler, C.V, Petersson, N.M, Iwai, H, Niemelae, M.A, Brohus, M, Overgaard, M.T. | | Deposit date: | 2022-10-18 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Allosteric changes in protein stability and dynamics as pathogenic mechanism for calmodulin variants not affecting Ca 2+ coordinating residues.
Cell Calcium, 117, 2023
|
|
