4WOL
 
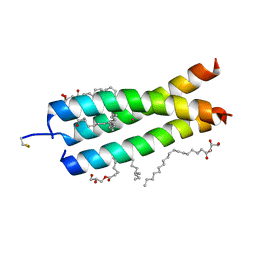 | | Crystal Structure of the DAP12 transmembrane domain in lipidic cubic phase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, POTASSIUM ION, TYRO protein tyrosine kinase-binding protein | | Authors: | Call, M.J, Call, M.E, Knoblich, K. | | Deposit date: | 2014-10-16 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Transmembrane Complexes of DAP12 Crystallized in Lipid Membranes Provide Insights into Control of Oligomerization in Immunoreceptor Assembly.
Cell Rep, 11, 2015
|
|
7XMN
 
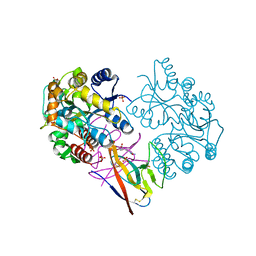 | | Structure of SARS-CoV-2 ORF8 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltodextrin-binding protein, ... | | Authors: | Chen, X, Xu, W. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycosylated, Lipid-Binding, CDR-Like Domains of SARS-CoV-2 ORF8 Indicate Unique Sites of Immune Regulation.
Microbiol Spectr, 11, 2023
|
|
5FSC
 
 | | Structure of tectonin 2 from Laccaria bicolor in complex with allyl- alpha_4-methyl-mannoside | | Descriptor: | MAGNESIUM ION, TECTONIN 2, prop-2-en-1-yl 4-O-methyl-alpha-D-mannopyranoside | | Authors: | Sommer, R, Bleuer, S, Titz, A, Kunzler, M, Varrot, A. | | Deposit date: | 2016-01-04 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Fungal Tectonin in Complex with O-Methylated Glycans Suggest Key Role in Innate Immune Defense.
Structure, 26, 2018
|
|
2X9B
 
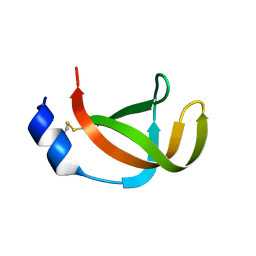 | | The filamentous phages fd and IF1 use different infection mechanisms | | Descriptor: | ATTACHMENT PROTEIN G3P | | Authors: | Lorenz, S.H, Jakob, R.P, Weininger, U, Dobbek, H, Schmid, F.X. | | Deposit date: | 2010-03-15 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | The Filamentous Phages Fd and If1 Use Different Mechanisms to Infect Escherichia Coli.
J.Mol.Biol., 405, 2011
|
|
5N9U
 
 | |
4Y9W
 
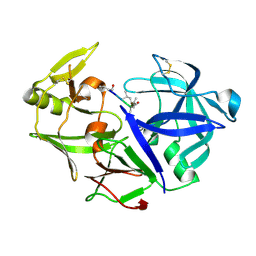 | | Aspartic Proteinase Sapp2 Secreted from Candida Parapsilosis at 0.82 A Resolution. | | Descriptor: | Aspartic acid endopeptidase Sapp2, PEPTIDE | | Authors: | Dostal, J, Brynda, J, Hruskova-Heidingsfeldova, O, Rezacova, P, Mareckova, L, Pichova, I. | | Deposit date: | 2015-02-17 | | Release date: | 2016-11-30 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | Atomic resolution crystal structure of Sapp2p, a secreted aspartic protease from Candida parapsilosis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6TPW
 
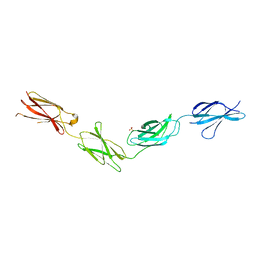 | | Crystal structures of FNIII domain one through four of the human leucocyte common antigen-related protein ( LAR) | | Descriptor: | Receptor-type tyrosine-protein phosphatase F, SULFATE ION | | Authors: | Vilstrup, J.P, Thirup, S.S, Simonsen, A, Birkefeldt, T, Strandbygaard, D. | | Deposit date: | 2019-12-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal and solution structures of fragments of the human leucocyte common antigen-related protein.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7BUT
 
 | |
2WN9
 
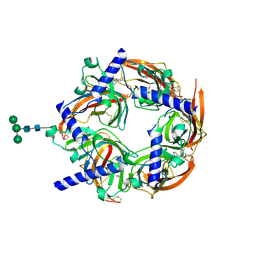 | | Crystal structure of Aplysia ACHBP in complex with 4-0H-DMXBA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(E)-5,6-DIHYDRO-2,3'-BIPYRIDIN-3(4H)-YLIDENEMETHYL]-3-METHOXYPHENOL, SOLUBLE ACETYLCHOLINE RECEPTOR, ... | | Authors: | Sulzenbacher, G, Hibbs, R, Shi, J, Talley, T, Conrod, S, Kem, W, Taylor, P, Marchot, P, Bourne, Y. | | Deposit date: | 2009-07-07 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural determinants for interaction of partial agonists with acetylcholine binding protein and neuronal alpha7 nicotinic acetylcholine receptor.
Embo J., 28, 2009
|
|
6TTW
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 4 (ASI_M3M_047) | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)-~{N}-piperidin-4-yl-oxolane-2-carboxamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
4YMG
 
 | | Crystal structure of SAM-bound Podospora anserina methyltransferase PaMTH1 | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Putative SAM-dependent O-methyltranferase, ... | | Authors: | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
7B81
 
 | |
2EGP
 
 | |
5LZS
 
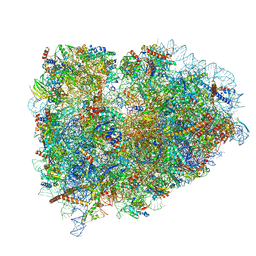 | | Structure of the mammalian ribosomal elongation complex with aminoacyl-tRNA, eEF1A, and didemnin B | | Descriptor: | (2~{S})-~{N}-[(2~{R})-1-[[(3~{S},6~{S},8~{S},12~{S},13~{R},16~{S},17~{R},20~{S},23~{S})-13-[(2~{S})-butan-2-yl]-20-[(4-methoxyphenyl)methyl]-6,17,21-trimethyl-3-(2-methylpropyl)-12-oxidanyl-2,5,7,10,15,19,22-heptakis(oxidanylidene)-8-propan-2-yl-9,18-dioxa-1,4,14,21-tetrazabicyclo[21.3.0]hexacosan-16-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-~{N}-methyl-1-[(2~{S})-2-oxidanylpropanoyl]pyrrolidine-2-carboxamide, 18S ribosomal RNA, 28S ribosomal RNA, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
7XBJ
 
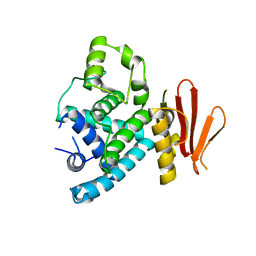 | | Txp40, an insecticidal toxin protein from Xenorhabdus nematophila | | Descriptor: | 40kDa insecticidal toxin | | Authors: | Kinkar, O, Kumar, A, Prashar, A, Hire, R.S, Makde, R.D. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The crystal structure of insecticidal protein Txp40 from Xenorhabdus nematophila reveals a two-domain unique binary toxin with homology to the toxin-antitoxin (TA) system.
Insect Biochem.Mol.Biol., 164, 2023
|
|
4YRA
 
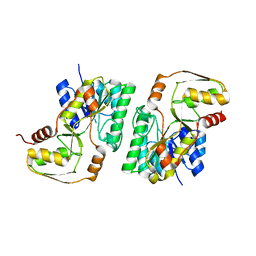 | | mouse TDH in the apo form | | Descriptor: | L-threonine 3-dehydrogenase, mitochondrial | | Authors: | He, C, Li, F. | | Deposit date: | 2015-03-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights on mouse l-threonine dehydrogenase: A regulatory role of Arg180 in catalysis
J.Struct.Biol., 192, 2015
|
|
6TPU
 
 | | Crystal structures of FNIII domain three and four of the human leucocyte common antigen-related protein, LAR | | Descriptor: | CHLORIDE ION, Receptor-type tyrosine-protein phosphatase F | | Authors: | Vilstrup, J.P, Thirup, S.S, Simonsen, A, Birkefeldt, T, Strandbygaard, D. | | Deposit date: | 2019-12-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal and solution structures of fragments of the human leucocyte common antigen-related protein.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5G6U
 
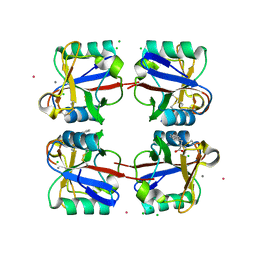 | | Crystal structure of langerin carbohydrate recognition domain with GlcNS6S | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Porkolab, V, Chabrol, E, Varga, N, Ordanini, S, Sutkeviciute, I, Thepaut, M, Bernardi, A, Fieschi, F. | | Deposit date: | 2016-07-21 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Rational-Differential Design of Highly Specific Glycomimetic Ligands: Targeting DC-SIGN and Excluding Langerin Recognition.
ACS Chem. Biol., 13, 2018
|
|
4YWK
 
 | |
5ZOJ
 
 | | Crystal structure of human SMAD2-MAN1 complex | | Descriptor: | Inner nuclear membrane protein Man1, Mothers against decapentaplegic homolog 2 | | Authors: | Miyazono, K, Ohno, Y, Ito, T, Tanokura, M. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Structural basis for receptor-regulated SMAD recognition by MAN1
Nucleic Acids Res., 46, 2018
|
|
6TTT
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 2 (ASI_M3M_140) | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-~{N}-methyl-3,4-bis(oxidanyl)oxolane-2-carboxamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
1PVV
 
 | |
1NP9
 
 | |
3PB2
 
 | |
3K9P
 
 | | The crystal structure of E2-25K and ubiquitin complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 K | | Authors: | Kang, G.B, Ko, S, Song, S.M, Lee, W, Eom, S.H. | | Deposit date: | 2009-10-16 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of E2-25K/UBB+1 interaction leading to proteasome inhibition and neurotoxicity
J.Biol.Chem., 285, 2010
|
|
