4ARI
 
 | | Ternary complex of E. coli leucyl-tRNA synthetase, tRNA(leu) and the benzoxaborole AN2679 in the editing conformation | | Descriptor: | GLYCEROL, LEUCINE--TRNA LIGASE, MAGNESIUM ION, ... | | Authors: | Palencia, A, Crepin, T, Vu, M.T, Lincecum Jr, T.L, Martinis, S.A, Cusack, S. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Dynamics of the Aminoacylation and Proofreading Functional Cycle of Bacterial Leucyl-tRNA Synthetase
Nat.Struct.Mol.Biol., 19, 2012
|
|
1RF9
 
 | | Crystal structure of cytochrome P450-cam with a fluorescent probe D-4-AD (Adamantane-1-carboxylic acid-5-dimethylamino-naphthalene-1-sulfonylamino-butyl-amide) | | Descriptor: | 1,2-ETHANEDIOL, ADAMANTANE-1-CARBOXYLIC ACID-5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONYLAMINO-BUTYL-AMIDE, Cytochrome P450-cam, ... | | Authors: | Hays, A.-M.A, Dunn, A.R, Gray, H.B, Stout, C.D, Goodin, D.B. | | Deposit date: | 2003-11-07 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational States of Cytochrome P450cam Revealed by Trapping of synthetic Molecular Wires
J.Mol.Biol., 344, 2004
|
|
4OT3
 
 | | Crystal structure of the S505L mutant of TAL effector dHax3 | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*AP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3 | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
3LWU
 
 | |
4B0J
 
 | | Crystal Structure of 3-hydroxydecanoyl-Acyl Carrier Protein Dehydratase (FabA) from Pseudomonas aeruginosa in complex with 5-(2- thienyl)-3-isoxazolyl methanol | | Descriptor: | (5-thiophen-2-ylisoxazol-3-yl)methanol, 3-HYDROXYDECANOYL-[ACYL-CARRIER-PROTEIN] DEHYDRATASE | | Authors: | Moynie, L, McMahon, S.A, Duthie, F.G, Naismith, J.H. | | Deposit date: | 2012-07-02 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights Into the Mechanism and Inhibition of the Beta-Hydroxydecanoyl-Acyl Carrier Protein Dehydratase from Pseudomonas Aeruginosa
J.Mol.Biol., 425, 2013
|
|
3LX0
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS D21N at cryogenic temperature | | Descriptor: | PHOSPHATE ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Doctrow, B.M, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-02-24 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cooperative Proton Binding in a Cluster of Carboxylic Residues
To be Published
|
|
5LEX
 
 | | Native human 20S proteasome in Mg-Acetate at 2.2 Angstrom | | Descriptor: | MAGNESIUM ION, PENTAETHYLENE GLYCOL, POTASSIUM ION, ... | | Authors: | Schrader, J, Henneberg, F, Mata, R, Tittmann, K, Schneider, T.R, Stark, H, Bourenkov, G, Chari, A. | | Deposit date: | 2016-06-30 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design.
Science, 353, 2016
|
|
3LXV
 
 | | Tyrosine 447 of Protocatechuate 3,4-Dioxygenase Controls Efficient Progress Through Catalysis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-NITROCATECHOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Purpero, V.M, Lipscomb, J.D. | | Deposit date: | 2010-02-25 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tyrosine 447 of Protocatechuate 3,4-Dioxygenase Controls Efficient Progress Through Catalysis
To be Published, 2010
|
|
4AXN
 
 | | Hallmarks of processive and non-processive glycoside hydrolases revealed from computational and crystallographic studies of the Serratia marcescens chitinases | | Descriptor: | ACETATE ION, CALCIUM ION, CHITINASE C1 | | Authors: | Payne, C.M, Baban, J, Synstad, B, Backe, P.H, Arvai, A.S, Dalhus, B, Bjoras, M, Eijsink, V.G.H, Sorlie, M, Beckham, G.T, Vaaje-Kolstad, G. | | Deposit date: | 2012-06-13 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Hallmarks of Processivity in Glycoside Hydrolases from Crystallographic and Computational Studies of the Serratia Marcescens Chitinases.
J.Biol.Chem., 287, 2012
|
|
3LTV
 
 | | Mouse-human sod1 chimera | | Descriptor: | Superoxide dismutase [Cu-Zn],Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Seetharaman, S.V, Taylor, A.B, Hart, P.J. | | Deposit date: | 2010-02-16 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | Structures of mouse SOD1 and human/mouse SOD1 chimeras.
Arch.Biochem.Biophys., 503, 2010
|
|
3LUO
 
 | | Crystal Structure and functional characterization of the thermophilic prolyl isomerase and chaperone SlyD | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, Suc-Ala-Leu-Pro-Phe-pNA, ZINC ION | | Authors: | Loew, C, Neumann, P, Weininger, U, Stubbs, M.T, Balbach, J. | | Deposit date: | 2010-02-18 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure Determination and Functional Characterization of the Metallochaperone SlyD from Thermus thermophilus
J.Mol.Biol., 398, 2010
|
|
3LUE
 
 | | Model of alpha-actinin CH1 bound to F-actin | | Descriptor: | Actin, cytoplasmic 1, Alpha-actinin-3 | | Authors: | Galkin, V.E, Orlova, A, Salmazo, A, Djinovic-Carugo, K, Egelman, E.H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Opening of tandem calponin homology domains regulates their affinity for F-actin.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4OO1
 
 | |
3LYC
 
 | |
5LUE
 
 | | Minor form of the recombinant cytotoxin-1 from N. oxiana | | Descriptor: | VC-1=CYTOTOXIN | | Authors: | Dubovskii, P.V, Dubinnyi, M.A, Shulepko, M.A, Lyukmanova, E.N, Dolgikh, D.A, Kirpichnikov, M.P, Efremov, R.G. | | Deposit date: | 2016-09-08 | | Release date: | 2017-09-20 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic "Portraits" of Recombinant and Native Cytotoxin I from Naja oxiana: How Close Are They?
Biochemistry, 56, 2017
|
|
3LVH
 
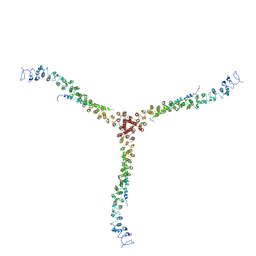 | | Crystal structure of a clathrin heavy chain and clathrin light chain complex | | Descriptor: | Clathrin heavy chain 1, Clathrin light chain B | | Authors: | Wilbur, J.D, Hwang, P.K, Ybe, J.A, Lane, M, Sellers, B.D, Jacobson, M.P, Fletterick, R.J, Brodsky, F.M. | | Deposit date: | 2010-02-20 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (9 Å) | | Cite: | Conformation switching of clathrin light chain regulates clathrin lattice assembly.
Dev.Cell, 18, 2010
|
|
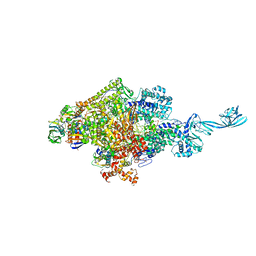
4G7O
 
 | |
4BHI
 
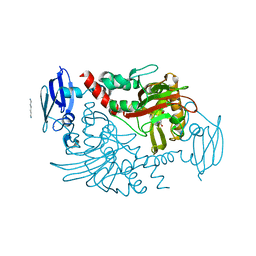 | | Three dimensional structure of human gamma-butyrobetaine hydroxylase in complex with 3-(1,1,1,2-Tetramethylhydrazin-1-ium-2-yl)propanoate | | Descriptor: | 3-(1,1,1,2-TETRAMETHYLHYDRAZIN-1-IUM-2-YL)PROPANOATE, GAMMA-BUTYROBETAINE DIOXYGENASE, HEXANE-1,6-DIAMINE, ... | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2013-04-03 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Targeting Carnitine Biosynthesis: Discovery of New Inhibitors Against Gamma-Butyrobetaine Hydroxylase.
J.Med.Chem., 57, 2014
|
|
4BDB
 
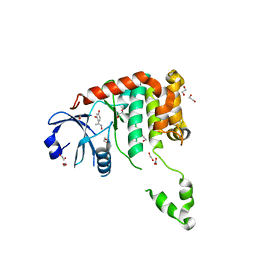 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(E)-C-methyl-N-oxidanyl-carbonimidoyl]benzene-1,3-diol, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
1R7A
 
 | | Sucrose Phosphorylase from Bifidobacterium adolescentis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, sucrose phosphorylase | | Authors: | Sprogoe, D, van den Broek, L.A.M, Mirza, O, Kastrup, J.S, Voragen, A.G.J, Gajhede, M, Skov, L.K. | | Deposit date: | 2003-10-21 | | Release date: | 2004-02-10 | | Last modified: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of sucrose phosphorylase from Bifidobacterium adolescentis.
Biochemistry, 43, 2004
|
|
3I9V
 
 | | Crystal structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus, oxidized, 2 mol/ASU | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Sazanov, L.A, Berrisford, J.M. | | Deposit date: | 2009-07-13 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the mechanism of respiratory complex I
J.Biol.Chem., 284, 2009
|
|
4GE1
 
 | | Structure of the tryptamine complex of the amine binding protein of Rhodnius prolixus | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, Biogenic amine-binding protein, GLYCEROL | | Authors: | Andersen, J.F, Chang, B.W, Xu, X, Mans, B.J, Ribeiro, J.M. | | Deposit date: | 2012-08-01 | | Release date: | 2013-01-02 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and ligand-binding properties of the biogenic amine-binding protein from the saliva of a blood-feeding insect vector of Trypanosoma cruzi.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4ASK
 
 | | CRYSTAL STRUCTURE OF JMJD3 WITH GSK-J1 | | Descriptor: | 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, COBALT (II) ION, LYSINE-SPECIFIC DEMETHYLASE 6B, ... | | Authors: | Chung, C, Mosley, J, Liddle, J. | | Deposit date: | 2012-05-01 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A Selective Jumonji H3K27 Demethylase Inhibitor Modulates the Proinflammatory Macrophage Response
Nature, 488, 2012
|
|
3LY7
 
 | |
4AVZ
 
 | | Tailspike protein mutant E372Q of E. coli bacteriophage HK620 | | Descriptor: | TAIL SPIKE PROTEIN, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM | | Authors: | Gohlke, U, Broeker, N.K, Mueller, J.J, Uetrecht, C, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2012-05-30 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Single Amino Acid Exchange in Bacteriophage Hk620 Tailspike Protein Results in Thousand-Fold Increase of its Oligosaccharide Affinity.
Glycobiology, 23, 2013
|
|
