7TIN
 
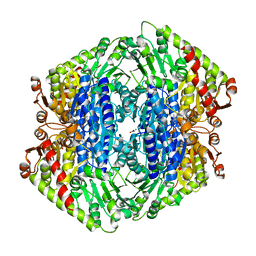 | | The Structure of S. aureus MenD | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Johnston, J.M, Stanborough, T, Ho, N.A.T, Akazong, E.W, Jiao, W. | | Deposit date: | 2022-01-14 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Allosteric inhibition of Staphylococcus aureus MenD by 1,4-dihydroxy naphthoic acid: a feedback inhibition mechanism of the menaquinone biosynthesis pathway.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 378, 2023
|
|
8WF5
 
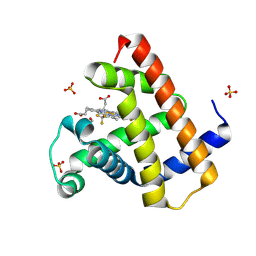 | | Horse heart myoglobin reconstituted with an iron complex of porphyrin bearing two CF3 groups (rMb(FePor(CF3)2)) | | Descriptor: | GLYCEROL, Myoglobin, PROTOPORPHYRIN(CF3)2 CONTAINING FE, ... | | Authors: | Kagawa, Y, Oohora, K, Himiyama, T, Suzuki, A, Hayashi, T. | | Deposit date: | 2023-09-19 | | Release date: | 2024-06-12 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Redox Engineering of Myoglobin by Cofactor Substitution to Enhance Cyclopropanation Reactivity.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
5V3I
 
 | |
5FSB
 
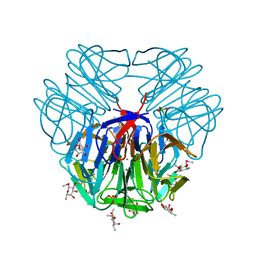 | | Structure of tectonin 2 from laccaria bicolor in complex with 2-o-methyl-methyl-seleno-beta-l-fucopyranoside | | Descriptor: | BORIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Sommer, R, Bleuer, S, Titz, A, Kunzler, M, Varrot, A. | | Deposit date: | 2016-01-04 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of Fungal Tectonin in Complex with O-Methylated Glycans Suggest Key Role in Innate Immune Defense.
Structure, 26, 2018
|
|
7NMY
 
 | | Crystal structure of beta-2-microglobulin D76Y mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NN5
 
 | | Crystal structure of beta-2-microglobulin D76K mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-24 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.242 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TVL
 
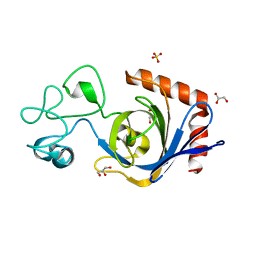 | | Viral AMG chitosanase V-Csn, apo structure | | Descriptor: | GLYCEROL, SULFATE ION, Viral chitosanase V-Csn | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
5E91
 
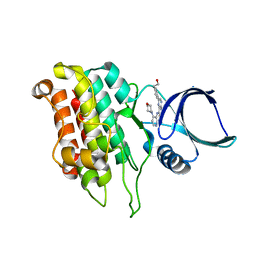 | |
5V4K
 
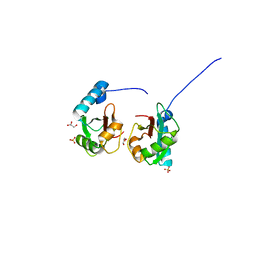 | | Crystal structure of NEDD4 LIR-fused human LC3B_2-119 | | Descriptor: | GLYCEROL, Microtubule-associated proteins 1A/1B light chain 3B,Microtubule-associated proteins 1A/1B light chain 3B,Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Qiu, Y, Zheng, Y, Schulman, B. | | Deposit date: | 2017-03-09 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Insights into links between autophagy and the ubiquitin system from the structure of LC3B bound to the LIR motif from the E3 ligase NEDD4.
Protein Sci., 26, 2017
|
|
7TVN
 
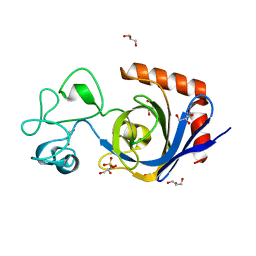 | | Viral AMG chitosanase V-Csn, D148N mutant | | Descriptor: | GLYCEROL, SULFATE ION, Viral chitosanase V-Csn D148N mutant | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
8PW2
 
 | |
5V4T
 
 | |
6NU9
 
 | |
8VZ0
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-400 | | Descriptor: | (1S,2R,4S)-N-(cyclopropylmethyl)-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-09 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7TLO
 
 | |
5EA2
 
 | | Crystal Structure of Holo NAD(P)H dehydrogenase, quinone 1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Pidugu, L.S, Mbimba, J.E, Ahmad, M, Pozharski, E, Sausville, E.A, Emadi, A, Toth, E.A. | | Deposit date: | 2015-10-15 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A direct interaction between NQO1 and a chemotherapeutic dimeric naphthoquinone.
Bmc Struct.Biol., 16, 2016
|
|
7TVO
 
 | | Viral AMG chitosanase V-Csn, E157Q mutant | | Descriptor: | GLYCEROL, SULFATE ION, Viral chitosanase V-Csn E157Q mutant | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
8P8T
 
 | | Ex vivo Ym2 crystal structure | | Descriptor: | Chitinase-like protein 4, GLYCEROL | | Authors: | Verschueren, K.H.G, Verstraete, K, Heyndrickx, I, Smole, U, Aegerter, H, Savvides, S.N, Lambrecht, B.N. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Ym1 protein crystals promote type 2 immunity.
Elife, 12, 2024
|
|
8VZ1
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-409 | | Descriptor: | 4,4'-[(1S,4S,5R)-5-(6-methoxy-3,4-dihydroquinoline-1(2H)-sulfonyl)-7-oxabicyclo[2.2.1]hept-2-ene-2,3-diyl]diphenol, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-09 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7MYT
 
 | |
6NGS
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(5-(3-(dimethylamino)propyl)-2,3-difluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{5-[3-(dimethylamino)propyl]-2,3-difluorophenyl}ethyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
5EAI
 
 | | Crystal Structure of NAD(P)H dehydrogenase, quinone 1 complexed with a chemotherapeutic naphthoquinone E6a | | Descriptor: | (2~{R},3~{R})-2-[(2~{S},3~{S})-3-bromanyl-1,4-bis(oxidanylidene)-2,3-dihydronaphthalen-2-yl]-3-oxidanyl-2,3-dihydronaphthalene-1,4-dione, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Pidugu, L.S, Mbimba, J.E, Ahmad, M, Pozharski, E, Sausville, E.A, Emadi, A, Toth, E.A. | | Deposit date: | 2015-10-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A direct interaction between NQO1 and a chemotherapeutic dimeric naphthoquinone.
Bmc Struct.Biol., 16, 2016
|
|
8P8Q
 
 | | Recombinant Ym1 crystal structure | | Descriptor: | ACETATE ION, Chitinase-like protein 3, GLYCEROL | | Authors: | Verschueren, K.H.G, Verstraete, K, Heyndrickx, I, Aegerter, H, Smole, U, Savvides, S.N, Lambrecht, B.N. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Ym1 protein crystals promote type 2 immunity.
Elife, 12, 2024
|
|
6NH4
 
 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 6-(3-fluoro-5-(2-((2R,4S)-4-fluoropyrrolidin-2-yl)ethyl)phenethyl)-4-methylpyridin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[2-(3-fluoro-5-{2-[(2R,4S)-4-fluoropyrrolidin-2-yl]ethyl}phenyl)ethyl]-4-methylpyridin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
7U3E
 
 | | GID4 in complex with compound 1 | | Descriptor: | Glucose-induced degradation protein 4 homolog, tert-butyl (1S,4S)-2,5-diazabicyclo[2.2.1]heptane-2-carboxylate | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
