7RVP
 
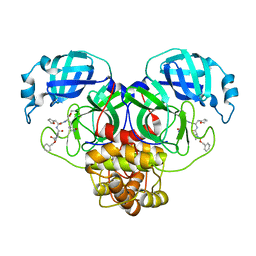 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI14 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-3-furan-2-yl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7VK0
 
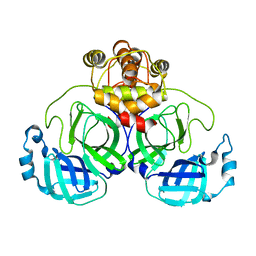 | |
1AC8
 
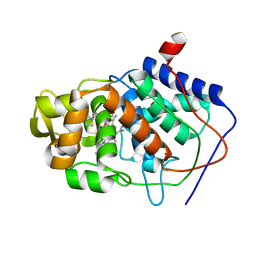 | | VARIATION IN THE STRENGTH OF A CH TO O HYDROGEN BOND IN AN ARTIFICIAL PROTEIN CAVITY (3,4,5-TRIMETHYLTHIAZOLE) | | Descriptor: | 3,4,5-TRIMETHYL-1,3-THIAZOLE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Bunte, S.W, Rosenfeld, R, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-14 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Variation in strength of an unconventional C-H to O hydrogen bond in an engineered protein cavity
J.Am.Chem.Soc., 119, 1997
|
|
7DKH
 
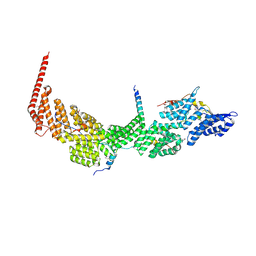 | | Crystal structure of the Ctr9/Paf1/Cdc73/Rtf1 quaternary complex | | Descriptor: | Cell division control protein 73, RNA polymerase II-associated protein 1, RNA polymerase-associated protein CTR9, ... | | Authors: | Chen, F.L, Liu, B.B, Guo, L, Li, D.F, Zhou, H, Long, J.F. | | Deposit date: | 2020-11-24 | | Release date: | 2021-11-24 | | Last modified: | 2022-01-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Core Module of the Yeast Paf1 Complex.
J.Mol.Biol., 434, 2021
|
|
5KGT
 
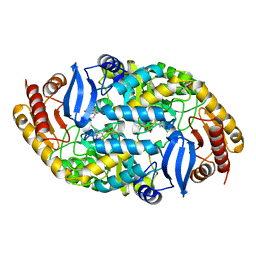 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, complexed with an inhibitor optimized from HTS lead: 1-[4-[4-(3-chlorophenyl)carbonylpiperidin-1-yl]phenyl]ethanone | | Descriptor: | 1-[4-[4-(3-chlorophenyl)carbonylpiperidin-1-yl]phenyl]ethanone, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Maize, K.M, Divakaran, A, Aldrich, C.C, Finzel, B.C. | | Deposit date: | 2016-06-13 | | Release date: | 2017-06-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Optimization of Pyridoxal 5'-Phosphate-Dependent Transaminase Enzyme (BioA) Inhibitors that Target Biotin Biosynthesis in Mycobacterium tuberculosis.
J. Med. Chem., 60, 2017
|
|
4ZJV
 
 | |
1AEG
 
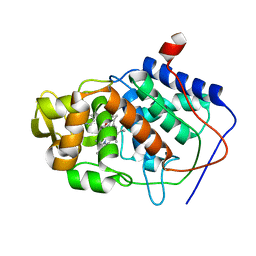 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (4-AMINOPYRIDINE) | | Descriptor: | 4-AMINOPYRIDINE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-24 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
1AEM
 
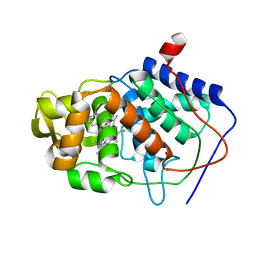 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (IMIDAZO[1,2-A]PYRIDINE) | | Descriptor: | CYTOCHROME C PEROXIDASE, IMIDAZO[1,2-A]PYRIDINE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-25 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
7S6W
 
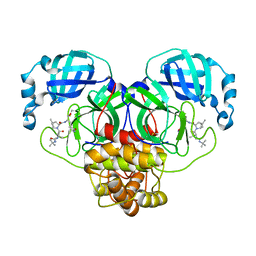 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI29 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S74
 
 | |
7V9I
 
 | | The Monomer mutant of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*T)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
4ZNT
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 3-Bromo-substituted OBHS derivative | | Descriptor: | 3-bromophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
1A88
 
 | | CHLOROPEROXIDASE L | | Descriptor: | CHLOROPEROXIDASE L | | Authors: | Hofmann, B, Toelzer, S, Pelletier, I, Altenbuchner, J, Van Pee, K.-H, Hecht, H.-J. | | Deposit date: | 1998-04-03 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural investigation of the cofactor-free chloroperoxidases.
J.Mol.Biol., 279, 1998
|
|
7JP4
 
 | |
167L
 
 | |
180L
 
 | |
4WP0
 
 | | Crystal structure of human kynurenine aminotransferase-I with a C-terminal V5-hexahistidine tag | | Descriptor: | Kynurenine--oxoglutarate transaminase 1 | | Authors: | Nadvi, N.A, Salam, N.K, Park, J, Akladios, F.N, Kapoor, V, Collyer, C.A, Gorrell, M.D, Church, W.B. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human kynurenine aminotransferase-I in a novel space group
To be published
|
|
1A3I
 
 | | X-RAY CRYSTALLOGRAPHIC DETERMINATION OF A COLLAGEN-LIKE PEPTIDE WITH THE REPEATING SEQUENCE (PRO-PRO-GLY) | | Descriptor: | ACETIC ACID, COLLAGEN-LIKE PEPTIDE | | Authors: | Kramer, R.Z, Vitagliano, L, Bella, J, Berisio, R, Mazzarella, L, Brodsky, B, Zagari, A, Berman, H.M. | | Deposit date: | 1998-01-22 | | Release date: | 1998-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | X-ray crystallographic determination of a collagen-like peptide with the repeating sequence (Pro-Pro-Gly).
J.Mol.Biol., 280, 1998
|
|
8ERL
 
 | |
3V83
 
 | | The 2.1 angstrom crystal structure of diferric human transferrin | | Descriptor: | BICARBONATE ION, FE (III) ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Noinaj, N, Steere, A, Mason, A.B, Buchanan, S.K. | | Deposit date: | 2011-12-22 | | Release date: | 2012-02-15 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural basis for iron piracy by pathogenic Neisseria.
Nature, 483, 2012
|
|
5HOH
 
 | | RIBONUCLEASE T1 (ASN9ALA/THR93ALA DOUBLEMUTANT) COMPLEXED WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Langhorst, U, Loris, R, Denisov, V.P, Doumen, J, Roose, P, Maes, D, Halle, B, Steyaert, J. | | Deposit date: | 1998-09-14 | | Release date: | 1998-09-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissection of the structural and functional role of a conserved hydration site in RNase T1.
Protein Sci., 8, 1999
|
|
5XVZ
 
 | | CATPO mutant - H246W | | Descriptor: | CALCIUM ION, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase, ... | | Authors: | Yuzugullu Karakus, Y, Goc, G, Balci, S, Pearson, A.R, Yorke, B. | | Deposit date: | 2017-06-28 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of the site of oxidase substrate binding in Scytalidium thermophilum catalase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6EAS
 
 | | Co-crystal of pseudokinase DRIK1 (drought responsive inactive kinase 1) bound to ENMD-2076 | | Descriptor: | 6-(4-methylpiperazin-1-yl)-N-(5-methyl-1H-pyrazol-3-yl)-2-[(E)-2-phenylethenyl]pyrimidin-4-amine, GLYCEROL, drought responsive inactive kinase 1 | | Authors: | Aquino, B, Counago, R.M, Fala, A.M, Massirer, K.B, Elkins, J.M, Arruda, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-08-03 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Co-crystal of pseudokinase DRIK1 with ENMD-2076
To be Published
|
|
4ZNM
 
 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form) | | Descriptor: | C-domain type II peptide synthetase, CHLORIDE ION, SODIUM ION | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-04 | | Release date: | 2015-05-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form)
To Be Published
|
|
7A8Y
 
 | |
