5UX2
 
 | |
7PVG
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with isonicotinic acid | | Descriptor: | Cholinephosphate cytidylyltransferase, pyridine-4-carboxylic acid | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-02 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
6N78
 
 | | Structure of the human JAK1 kinase domain with compound 21 | | Descriptor: | GLYCEROL, N-{3-[5-chloro-2-(difluoromethoxy)phenyl]-1-methyl-1H-pyrazol-4-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
5DOW
 
 | |
8PZ7
 
 | | crystal structure of VDR complex with D-Bishomo-1a,25-dihydroxyvitamin D3 Analog 57 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(4~{a}~{R},5~{R},9~{a}~{S})-4~{a}-methyl-5-[(2~{R})-6-methyl-6-oxidanyl-heptan-2-yl]-3,4,5,8,9,9~{a}-hexahydro-2~{H}-benzo[7]annulen-1-ylidene]ethylidene]-2-methyl-cyclohexane-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2023-07-27 | | Release date: | 2023-08-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Design, synthesis, and biological activity of D-bishomo-1 alpha ,25-dihydroxyvitamin D 3 analogs and their crystal structures with the vitamin D nuclear receptor.
Eur.J.Med.Chem., 271, 2024
|
|
6N7A
 
 | | Structure of the human JAK1 kinase domain with compound 39 | | Descriptor: | GLYCEROL, N-[3-(5-chloro-2-methoxyphenyl)-1-methyl-1H-pyrazol-4-yl]-2-methyl-2H-pyrazolo[4,3-c]pyridine-7-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
7NAI
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with 3AD | | Descriptor: | Sterile alpha and TIR motif-containing protein 1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(8-azanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Shi, Y, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
8SM3
 
 | | Structure of Bacillus cereus VD045 Gabija GajA-GajB Complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB, SULFATE ION | | Authors: | Antine, S.P, Mooney, S.E, Johnson, A.G, Kranzusch, P.J. | | Deposit date: | 2023-04-25 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of Gabija anti-phage defence and viral immune evasion.
Nature, 625, 2024
|
|
5HGP
 
 | |
7PTG
 
 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with EBL2888 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1S)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
8SXS
 
 | |
7PJO
 
 | |
5VAL
 
 | | BRAF in Complex with N-(3-(tert-butyl)phenyl)-4-methyl-3-(6-morpholinopyrimidin-4-yl)benzamide | | Descriptor: | N-(3-tert-butylphenyl)-4-methyl-3-[6-(morpholin-4-yl)pyrimidin-4-yl]benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2017-03-27 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Design and Discovery of N-(2-Methyl-5'-morpholino-6'-((tetrahydro-2H-pyran-4-yl)oxy)-[3,3'-bipyridin]-5-yl)-3-(trifluoromethyl)benzamide (RAF709): A Potent, Selective, and Efficacious RAF Inhibitor Targeting RAS Mutant Cancers.
J. Med. Chem., 60, 2017
|
|
8SMF
 
 | | Structure of SPO1 phage Tad2 in complex with 1''-3' gcADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, Gp34.65, MAGNESIUM ION | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
5HH5
 
 | |
7W11
 
 | | glycosyltransferase | | Descriptor: | DIGITOXIGENIN, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7NNJ
 
 | | Crystal Structure of NUDT4 (Diphosphoinositol polyphosphate phosphohydrolase 2) in complex with 4-O-Bn-1-PCP-InsP4 (AMR2105) | | Descriptor: | 1,2-ETHANEDIOL, Diphosphoinositol polyphosphate phosphohydrolase 2, FORMIC ACID, ... | | Authors: | Dubianok, Y, Arruda Bezerra, G, Raux, B, Diaz Saez, L, Riley, A.M, Potter, B.V.L, Huber, K.V.M, von Delft, F. | | Deposit date: | 2021-02-24 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Crystal Structure of NUDT4 (Diphosphoinositol polyphosphate phosphohydrolase 2) in complex with 4-O-Bn-1-PCP-InsP4 (AMR2105)
To Be Published
|
|
7YN3
 
 | | Crystal structure of CcbD complex with CcbZ carrier protein domain | | Descriptor: | 1,1'-ethane-1,2-diyldipyrrolidine-2,5-dione, 4'-PHOSPHOPANTETHEINE, CcbD, ... | | Authors: | Mori, T, Lyu, S, Abe, I. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of CcbD complex with carrier protein
To Be Published
|
|
5VAS
 
 | | Pekin duck egg lysozyme isoform III (DEL-III), orthorhombic form | | Descriptor: | GLYCEROL, Lysozyme, PHOSPHATE ION | | Authors: | Christie, M, Christ, D, Langley, D.B. | | Deposit date: | 2017-03-27 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of antigen recognition: crystal structure of duck egg lysozyme.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
8PJM
 
 | |
7MUB
 
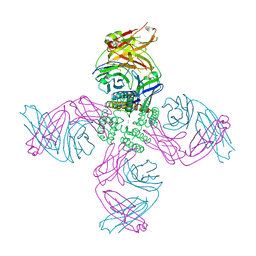 | | KcsA Open gate E71V mutant in Potassium | | Descriptor: | Fab heavy chain, Fab light chain, POTASSIUM ION, ... | | Authors: | Rohaim, A, Li, J, Weingarth, M, Roux, B. | | Deposit date: | 2021-05-14 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A distinct mechanism of C-type inactivation in the Kv-like KcsA mutant E71V.
Nat Commun, 13, 2022
|
|
7YOQ
 
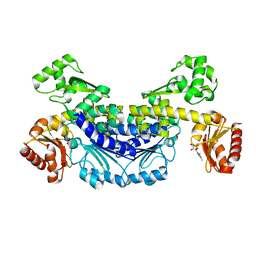 | | Recj2 and CMP complex from Methanocaldococcus jannaschii | | Descriptor: | [5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)furan-2-yl]methyl dihydrogen phosphate, nuclease RecJ2 | | Authors: | Wang, W.W, Liu, X.P. | | Deposit date: | 2022-08-02 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.265 Å) | | Cite: | The structure of nuclease RecJ2 from Methanocaldococcus jannaschii
To Be Published
|
|
7W09
 
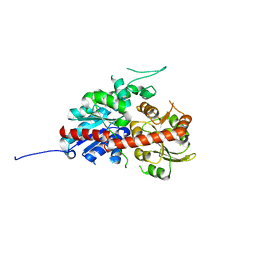 | | UGT74AN2, Plant Steroid Glycosyltransferase | | Descriptor: | Glycosyltransferase | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7W0Z
 
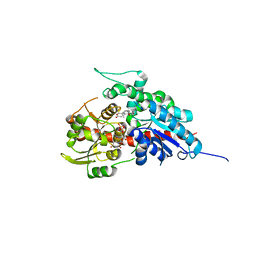 | | Glycosyltranferase UGT74AN2 | | Descriptor: | 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
8VZ4
 
 | |
