7M4U
 
 | | A. baumannii Ribosome-Eravacycline complex: 30S | | Descriptor: | 16s Ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
7M4Z
 
 | | A. baumannii Ribosome-Eravacycline complex: hpf-bound 70S | | Descriptor: | 16s Ribosomal RNA, 23s ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
3B2R
 
 | | Crystal Structure of PDE5A1 catalytic domain in complex with Vardenafil | | Descriptor: | 2-{2-ETHOXY-5-[(4-ETHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-5-METHYL-7-PROPYLIMIDAZO[5,1-F][1,2,4]TRIAZIN-4(1H)-ONE, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Huanchen, W, Mengchun, Y, Howard, R, Sharron, H.F, Hengming, K. | | Deposit date: | 2007-10-19 | | Release date: | 2008-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Conformational variations of both phosphodiesterase-5 and inhibitors provide the structural basis for the physiological effects of vardenafil and sildenafil.
Mol.Pharmacol., 73, 2008
|
|
7M4X
 
 | | A. baumannii Ribosome-Eravacycline complex: P-site tRNA 70S | | Descriptor: | 16s Ribosomal RNA, 23s ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
7M4W
 
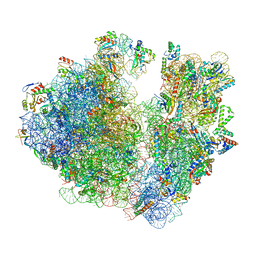 | | A. baumannii Ribosome-Eravacycline complex: Empty 70S | | Descriptor: | 16s Ribosomal RNA, 23s ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
7M4Y
 
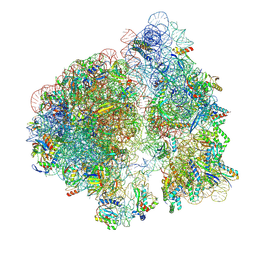 | | A. baumannii Ribosome-Eravacycline complex: E-site tRNA 70S | | Descriptor: | 16s Ribosomal RNA, 23s ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
8A12
 
 | | Plasmodium falciparum Myosin A full-length, post-rigor state complexed to Mg.ATP-gamma-S | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Moussaoui, D, Robblee, J.P, Auguin, D, Fisher, F, Fagnant, P.M, MacFarlane, J.E, Mueller-Dieckmann, C, Baum, J, Robert-Paganin, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2022-05-31 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mechanism of small molecule inhibition of Plasmodium falciparum myosin A informs antimalarial drug design.
Nat Commun, 14, 2023
|
|
3IE3
 
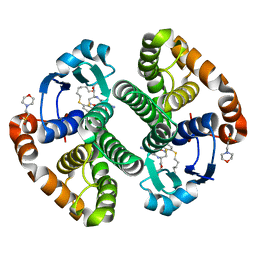 | | Structural basis for the binding of the anti-cancer compound 6-(7-Nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol (NBDHEX) to human glutathione S-transferases | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[(7-nitro-2,1,3-benzoxadiazol-4-yl)sulfanyl]hexan-1-ol, GLUTATHIONE, ... | | Authors: | Federici, L, Lo Sterzo, C, Di Matteo, A, Scaloni, F, Federici, G, Caccuri, A.M. | | Deposit date: | 2009-07-22 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the binding of the anticancer compound 6-(7-nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol to human glutathione s-transferases
Cancer Res., 69, 2009
|
|
3QUK
 
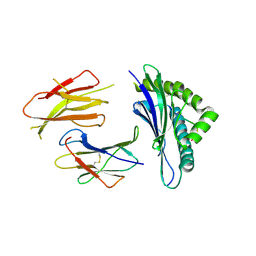 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 altered peptide ligand (Y4A) | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Allerbring, E, Duru, A.D, Uchtenhagen, H, Madhurantakam, C, Grimm, S, Tomek, M.B, Mazumdar, P.A, Spetz, A, Friemann, R, Sandalova, T, Uhlin, M, Nygren, P, Achour, A. | | Deposit date: | 2011-02-24 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Unexpected T-cell recognition of an altered peptide ligand is driven by reversed thermodynamics.
Eur.J.Immunol., 42, 2012
|
|
3QUL
 
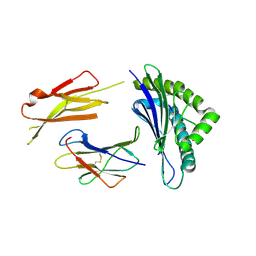 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 altered peptide ligand (Y4S) | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Allerbring, E, Duru, A.D, Uchtenhagen, H, Madhurantakam, C, Grimm, S, Tomek, M.B, Mazumdar, P.A, Spetz, A, Friemann, R, Sandalova, T, Uhlin, M, Nygren, P, Achour, A. | | Deposit date: | 2011-02-24 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected T-cell recognition of an altered peptide ligand is driven by reversed thermodynamics.
Eur.J.Immunol., 42, 2012
|
|
6W8S
 
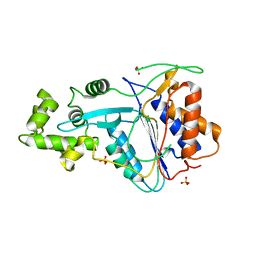 | | Crystal structure of metacaspase 4 from Arabidopsis | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.484 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
6W8T
 
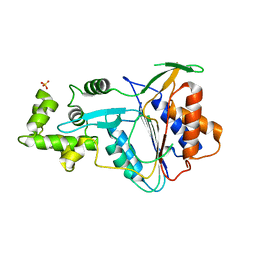 | | Crystal structure of metacaspase 4 from Arabidopsis (microcrystals treated with calcium) | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
7D8K
 
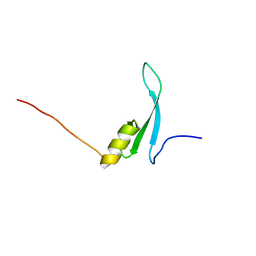 | | Solution structure of the methyl-CpG binding domain of MBD6 from Arabidopsis thaliana | | Descriptor: | Methyl-CpG-binding domain-containing protein 6 | | Authors: | Mahana, Y, Ohki, I, Walinda, E, Morimoto, D, Sugase, K, Shirakawa, M. | | Deposit date: | 2020-10-08 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into Methylated DNA Recognition by the Methyl-CpG Binding Domain of MBD6 from Arabidopsis thaliana .
Acs Omega, 7, 2022
|
|
7R47
 
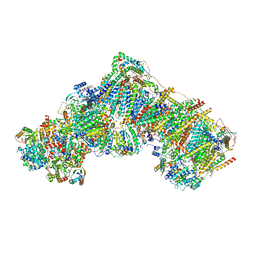 | | Bovine complex I in the presence of IM1761092, deactive class iii (Composite map) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-carbamimidoyl-3-[2-(3-chloranyl-4-iodanyl-phenyl)ethyl]guanidine, ... | | Authors: | Bridges, H.R, Blaza, J.N, Yin, Z, Chung, I, Hirst, J. | | Deposit date: | 2022-02-08 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis of mammalian respiratory complex I inhibition by medicinal biguanides.
Science, 379, 2023
|
|
6W8R
 
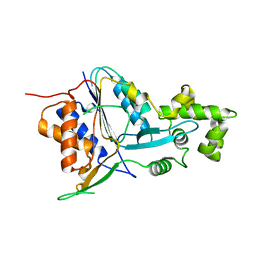 | | Crystal structure of metacaspase 4 C139A from Arabidopsis | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
5MPR
 
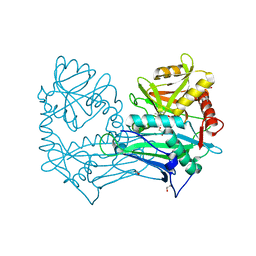 | | Single Amino Acid Variant of Human Mitochondrial Branched Chain Amino Acid Aminotransferase 2 | | Descriptor: | 1,2-ETHANEDIOL, Branched-chain-amino-acid aminotransferase, mitochondrial, ... | | Authors: | Hakansson, M, Walse, B, Nilsson, C, Anderson, L.C. | | Deposit date: | 2016-12-18 | | Release date: | 2017-07-19 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Intact Protein Analysis at 21 Tesla and X-Ray Crystallography Define Structural Differences in Single Amino Acid Variants of Human Mitochondrial Branched-Chain Amino Acid Aminotransferase 2 (BCAT2).
J. Am. Soc. Mass Spectrom., 28, 2017
|
|
8BRY
 
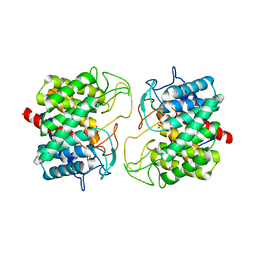 | | Room-temperature structure of Pedobacter heparinus N-acetylglucosamine 2-epimerase at atmospheric pressure | | Descriptor: | CHLORIDE ION, N-acylglucosamine 2-epimerase, PHOSPHATE ION | | Authors: | Lieske, J, Saouane, S, Assmann, M, Zaun, H, Kuballa, J, Meents, A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
8BS1
 
 | | Room-temperature structure of SARS-CoV-2 Main protease at atmospheric pressure | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Lieske, J, Saouane, S, Guenther, S, Reinke, P.Y.A, Rahmani Mashhour, A, Meents, A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
1H2R
 
 | |
1BP1
 
 | | CRYSTAL STRUCTURE OF BPI, THE HUMAN BACTERICIDAL PERMEABILITY-INCREASING PROTEIN | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERICIDAL/PERMEABILITY-INCREASING PROTEIN | | Authors: | Beamer, L.J, Carroll, S.F, Eisenberg, D. | | Deposit date: | 1997-04-08 | | Release date: | 1997-09-04 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human BPI and two bound phospholipids at 2.4 angstrom resolution.
Science, 276, 1997
|
|
1J52
 
 | |
6TTO
 
 | | N-terminally truncated hyoscyamine 6-hydroxylase (tH6H) in complex with 2-oxoglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, FORMIC ACID, ... | | Authors: | Kluza, A, Mrugala, B, Porebski, P.J, Kurpiewska, K, Niedzialkowska, E, Weiss, M.S, Borowski, T. | | Deposit date: | 2019-12-29 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Regioselectivity of hyoscyamine 6 beta-hydroxylase-catalysed hydroxylation as revealed by high-resolution structural information and QM/MM calculations.
Dalton Trans, 49, 2020
|
|
1TYP
 
 | |
6TTM
 
 | | Hyoscyamine 6-hydroxylase in complex with N-oxalylglycine and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Hyoscyamine 6 beta-hydroxylase, ... | | Authors: | Kluza, A, Kurpiewska, K, Mrugala, B, Porebski, P.J, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-12-29 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Regioselectivity of hyoscyamine 6 beta-hydroxylase-catalysed hydroxylation as revealed by high-resolution structural information and QM/MM calculations.
Dalton Trans, 49, 2020
|
|
6TTN
 
 | | N-terminally truncated hyoscyamine 6-hydroxylase (tH6H) in complex with N-oxalylglycine and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, Hyoscyamine 6 beta-hydroxylase, N-OXALYLGLYCINE, ... | | Authors: | Kluza, A, Mrugala, B, Porebski, P.J, Kurpiewska, K, Niedzialkowska, E, Weiss, M.S, Borowski, T. | | Deposit date: | 2019-12-29 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Regioselectivity of hyoscyamine 6 beta-hydroxylase-catalysed hydroxylation as revealed by high-resolution structural information and QM/MM calculations.
Dalton Trans, 49, 2020
|
|
