2BTJ
 
 | | Fluorescent Protein EosFP - red form | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP | | Authors: | Nar, H, Nienhaus, K, Wiedenmann, J, Nienhaus, G.U. | | Deposit date: | 2005-06-01 | | Release date: | 2005-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Photo-Induced Protein Cleavage and Green-to-Red Conversion of Fluorescent Protein Eosfp.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6XHF
 
 | | Structure of Phenylalanyl-5'-O-adenosine phosphoramidate | | Descriptor: | (2~{S})-2-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]amino]-3-phenyl-propanoic acid, CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4FN3
 
 | |
4FKP
 
 | |
3UOI
 
 | |
8F4U
 
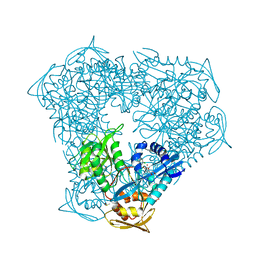 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with azelastine | | Descriptor: | 4-[(4-chlorophenyl)methyl]-2-[(4S)-1-methylazepan-4-yl]phthalazin-1(2H)-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
1I0R
 
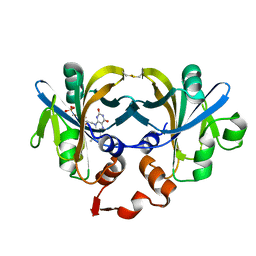 | | CRYSTAL STRUCTURE OF FERRIC REDUCTASE FROM ARCHAEOGLOBUS FULGIDUS | | Descriptor: | CONSERVED HYPOTHETICAL PROTEIN, FLAVIN MONONUCLEOTIDE | | Authors: | Chiu, H.-J, Johnson, E, Schroder, I, Rees, D.C. | | Deposit date: | 2001-01-29 | | Release date: | 2001-05-02 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of a novel ferric reductase from the hyperthermophilic archaeon Archaeoglobus fulgidus and its complex with NADP+.
Structure, 9, 2001
|
|
4FL0
 
 | | Crystal structure of ALD1 from Arabidopsis thaliana | | Descriptor: | Aminotransferase ALD1, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sobolev, V, Edelman, M, Dym, O, Unger, T, Albeck, S, Kirma, M, Galili, G, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2012-06-14 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of ALD1, a plant-specific homologue of the universal diaminopimelate aminotransferase enzyme of lysine biosynthesis.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3UQH
 
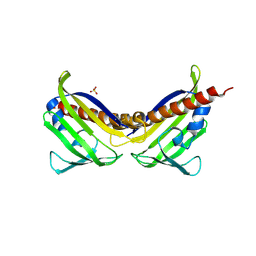 | | Crystal structure of aba receptor pyl10 (apo) | | Descriptor: | Abscisic acid receptor PYL10, SULFATE ION | | Authors: | Sun, D.M, Wang, H.P, Wu, M.H, Zang, J.Y, Tian, C.L. | | Deposit date: | 2011-11-20 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Aba Receptor Pyl10
To be Published
|
|
4FLK
 
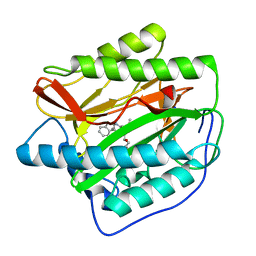 | | Human MetAP1 with bengamide analog Y10, in Mn form | | Descriptor: | (E,2R,3R,4S,5R)-N-(2,3-dihydro-1H-inden-2-yl)-2-methoxy-8,8-dimethyl-3,4,5-tris(oxidanyl)non-6-enamide, MANGANESE (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Ye, Q.Z, Xu, W. | | Deposit date: | 2012-06-14 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural analysis of bengamide derivatives as inhibitors of methionine aminopeptidases.
J.Med.Chem., 55, 2012
|
|
6XP8
 
 | |
4FNH
 
 | | Crystal structure of IsdI-W66Y in complex with heme | | Descriptor: | Heme-degrading monooxygenase isdI, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ukpabi, G.N, Murphy, M.E.P. | | Deposit date: | 2012-06-19 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inactivation of the heme degrading enzyme IsdI by an active site substitution that diminishes heme ruffling.
J.Biol.Chem., 287, 2012
|
|
3UJ3
 
 | |
3UR0
 
 | | Crystal structures of murine norovirus RNA-dependent RNA polymerase in complex with Suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFONIC ACID, RNA-dependent RNA polymerase, SULFATE ION | | Authors: | Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-05-02 | | Last modified: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Inhibition of Norovirus RNA-Dependent RNA Polymerases.
J.Mol.Biol., 419, 2012
|
|
6XNT
 
 | | Crystal structure of I91A mutant of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, octyl beta-D-glucopyranoside | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Bonsor, D, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2020-07-04 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Commun Biol, 4, 2021
|
|
4FNX
 
 | |
6XHD
 
 | | Structure of Prolinyl-5'-O-adenosine phosphoramidate | | Descriptor: | (2~{S})-1-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]pyrrolidine-2-carboxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3UJI
 
 | | Crystal structure of anti-HIV-1 V3 Fab 2558 in complex with MN peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein gp160, ... | | Authors: | Kong, X.P. | | Deposit date: | 2011-11-07 | | Release date: | 2011-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human Anti-V3 HIV-1 Monoclonal Antibodies Encoded by the VH5-51/VL Lambda Genes Define a Conserved Antigenic Structure.
Plos One, 6, 2011
|
|
4FN0
 
 | | Crystal structure of mouse nectin-2 extracellular fragment D1-D2, 2nd crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Poliovirus receptor-related protein 2, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2012-06-18 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Nectin ectodomain structures reveal a canonical adhesive interface.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4FO9
 
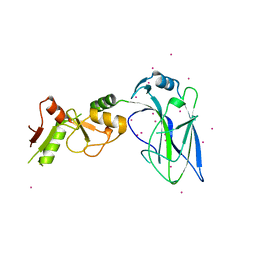 | | Crystal structure of the E3 SUMO Ligase PIAS2 | | Descriptor: | E3 SUMO-protein ligase PIAS2, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Dong, A, Hu, J, Dobrovetsky, E, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of the E3 SUMO Ligase PIAS2
to be published
|
|
6XNW
 
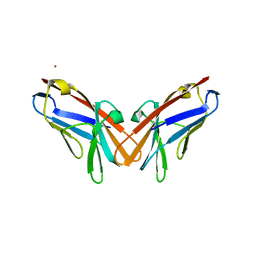 | | Crystal structure of V39A mutant of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, NICKEL (II) ION | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Bonsor, D, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2020-07-04 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Commun Biol, 4, 2021
|
|
4FOD
 
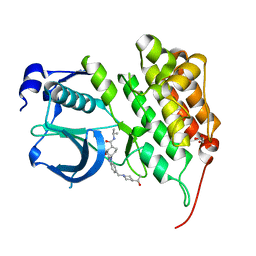 | | Crystal structure of human anaplastic lymphoma kinase in complex with acyliminobenzimidazole inhibitor 36 | | Descriptor: | 4-fluoro-N-{(2E)-6-{[4-(2-hydroxypropan-2-yl)piperidin-1-yl]methyl}-1-[cis-4-(propan-2-ylcarbamoyl)cyclohexyl]-1,3-dihydro-2H-benzimidazol-2-ylidene}benzamide, ALK tyrosine kinase receptor, GLYCEROL | | Authors: | Whittington, D.A, Epstein, L.F, Chen, H. | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Discovery and Optimization of a Novel Class of Potent, Selective, and Orally Bioavailable Anaplastic Lymphoma Kinase (ALK) Inhibitors with Potential Utility for the Treatment of Cancer.
J.Med.Chem., 55, 2012
|
|
4FO8
 
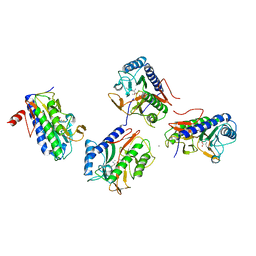 | | Pseudomonas aeruginosa MetAP with Met, in Mn form | | Descriptor: | MANGANESE (II) ION, METHIONINE, Methionine aminopeptidase | | Authors: | Ye, Q.Z, Lu, J.P. | | Deposit date: | 2012-06-20 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Methionine excision captured by the structures of a
methionine aminopeptidase
To be published
|
|
3UV7
 
 | | Ec_IspH in complex with buta-2,3-dienyl diphosphate (1300) | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER, buta-2,3-dien-1-yl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-29 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
4FOM
 
 | | Crystal structure of human nectin-3 full ectodomain (D1-D3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Poliovirus receptor-related protein 3, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Harrison, O.J, Jin, X, Brasch, J, Shapiro, L. | | Deposit date: | 2012-06-20 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.93 Å) | | Cite: | Nectin ectodomain structures reveal a canonical adhesive interface.
Nat.Struct.Mol.Biol., 19, 2012
|
|
