2G71
 
 | | Structure of hPNMT with inhibitor 3-fluoromethyl-7-trifluoropropyl-THIQ and AdoHcy | | Descriptor: | (3R)-3-(FLUOROMETHYL)-N-(3,3,3-TRIFLUOROPROPYL)-1,2,3,4-TETRAHYDROISOQUINOLINE-7-SULFONAMIDE, GLYCEROL, Phenylethanolamine N-methyltransferase, ... | | Authors: | Tyndall, J.D.A, Gee, C.L, Martin, J.L. | | Deposit date: | 2006-02-27 | | Release date: | 2007-02-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzyme Adaptation to Inhibitor Binding: A Cryptic Binding Site in Phenylethanolamine N-Methyltransferase
J.Med.Chem., 50, 2007
|
|
2QML
 
 | |
2GBV
 
 | | C6A/C111A/C57A/C146A holo CuZn Superoxide dismutase | | Descriptor: | COPPER (I) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Hornberg, A, Logan, D.T, Marklund, S.L, Oliveberg, M. | | Deposit date: | 2006-03-11 | | Release date: | 2007-01-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Coupling between Disulphide Status, Metallation and Dimer Interface Strength in Cu/Zn Superoxide Dismutase
J.Mol.Biol., 365, 2007
|
|
2GAM
 
 | | X-ray crystal structure of murine leukocyte-type Core 2 b1,6-N-acetylglucosaminyltransferase (C2GnT-L) in complex with Galb1,3GalNAc | | Descriptor: | beta-1,6-N-acetylglucosaminyltransferase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose | | Authors: | Pak, J.E, Rini, J.M. | | Deposit date: | 2006-03-09 | | Release date: | 2006-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray Crystal Structure of Leukocyte Type Core 2 beta1,6-N-Acetylglucosaminyltransferase: Evidence for a covergence of metal ion independent glycosyltransferase mechanism.
J.Biol.Chem., 281, 2006
|
|
2GBA
 
 | | Reduced Cu(I) form at pH 4 of P52G mutant of amicyanin | | Descriptor: | COPPER (I) ION, amicyanin | | Authors: | Ma, J.K, Carrell, C.J, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2006-03-10 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Site-Directed Mutagenesis of Proline 52 To Glycine in Amicyanin Converts a True Electron Transfer Reaction into One that Is Conformationally Gated.
Biochemistry, 45, 2006
|
|
2GEJ
 
 | | Crystal Structure of phosphatidylinositol mannosyltransferase (PimA) from Mycobacterium smegmatis in complex with GDP-Man | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, PHOSPHATIDYLINOSITOL MANNOSYLTRANSFERASE (PimA) | | Authors: | Guerin, M.E, Buschiazzo, A, Kordulakova, J, Jackson, M, Alzari, P.M. | | Deposit date: | 2006-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria.
J.Biol.Chem., 282, 2007
|
|
5JYT
 
 | |
2GGC
 
 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | COBALT (II) ION, METHIONINE, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
2GH2
 
 | |
2GHB
 
 | | Thermotoga maritima maltotriose binding protein, ligand free form | | Descriptor: | maltose ABC transporter, periplasmic maltose-binding protein | | Authors: | Cuneo, M.J, Changela, A, Hocker, B, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2006-03-27 | | Release date: | 2007-02-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | T. maritima maltotriose binding protein open form
To be Published
|
|
2GHM
 
 | | Mutated MAP kinase P38 (Mus Musculus) in complex with Inhbitor PG-895449 | | Descriptor: | 3-(2-CHLOROBENZYL)-1-(2-{[(1S)-2-HYDROXY-1,2-DIMETHYLPROPYL]AMINO}PYRIMIDIN-4-YL)-1-(4-METHOXYPHENYL)UREA, Mitogen-activated protein kinase 14 | | Authors: | Walter, R.L, Mekel, M.J, Evdokimov, A.G, Pokross, M.E, Maier, J.A. | | Deposit date: | 2006-03-27 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Development of N-2,4-pyrimidine-N-phenyl-N'-phenyl ureas as inhibitors of tumor necrosis factor alpha (TNF-alpha) synthesis. Part 1.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2GAI
 
 | |
4V95
 
 | | Crystal structure of YAEJ bound to the 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Gagnon, M.G, Seetharaman, S.V, Bulkley, D.P, Steitz, T.A. | | Deposit date: | 2012-01-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome.
Science, 335, 2012
|
|
2FP4
 
 | |
5RSL
 
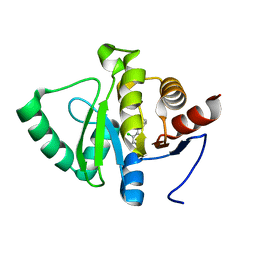 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000365052868 | | Descriptor: | 6-[(1s,4s)-2-azabicyclo[2.2.2]octan-2-yl]-5-chloropyrimidin-4-amine, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
4V4I
 
 | | Crystal Structure of a 70S Ribosome-tRNA Complex Reveals Functional Interactions and Rearrangements. | | Descriptor: | 16S SMALL SUBUNIT RIBOSOMAL RNA, 23S LARGE SUBUNIT RIBOSOMAL RNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Trakhanov, S, Laurberg, M, Noller, H.F. | | Deposit date: | 2007-02-15 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Crystal Structure of a 70S Ribosome-tRNA Complex Reveals Functional Interactions and Rearrangements
Cell(Cambridge,Mass.), 126, 2006
|
|
4V5Y
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with paromomycin and ribosome recycling factor (RRF). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-19 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.45 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4V7S
 
 | | Crystal structure of the E. coli ribosome bound to telithromycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-05 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2547 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V7T
 
 | | Crystal structure of the E. coli ribosome bound to chloramphenicol. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.1942 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5K2F
 
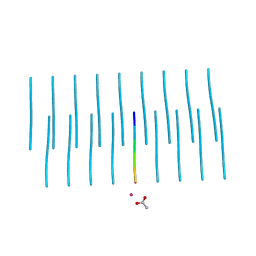 | | Structure of NNQQNY from yeast prion Sup35 with cadmium acetate determined by MicroED | | Descriptor: | ACETATE ION, CADMIUM ION, Eukaryotic peptide chain release factor GTP-binding subunit | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2FS2
 
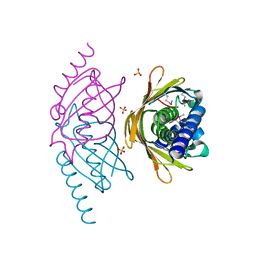 | | Structure of the E. coli PaaI protein from the phyenylacetic acid degradation operon | | Descriptor: | Phenylacetic acid degradation protein paaI, SULFATE ION | | Authors: | Kniewel, R, Buglino, J.A, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-01-20 | | Release date: | 2006-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, Function, and Mechanism of the Phenylacetate Pathway Hot Dog-fold Thioesterase PaaI
J.Biol.Chem., 281, 2006
|
|
2FS8
 
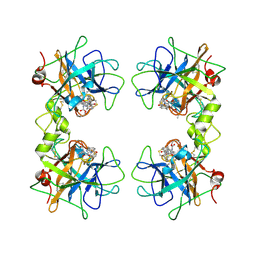 | | Human beta-tryptase II with inhibitor CRA-29382 | | Descriptor: | ALLYL {(1S)-1-[(5-{4-[(2,3-DIHYDRO-1H-INDEN-2-YLAMINO)CARBONYL]BENZYL}-1,2,4-OXADIAZOL-3-YL)CARBONYL]-3-PYRROLIDIN-3-YLPROPYL}CARBAMATE, Tryptase beta-2 | | Authors: | Somoza, J.R. | | Deposit date: | 2006-01-21 | | Release date: | 2006-03-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-guided design of Peptide-based tryptase inhibitors.
Biochemistry, 45, 2006
|
|
4V8E
 
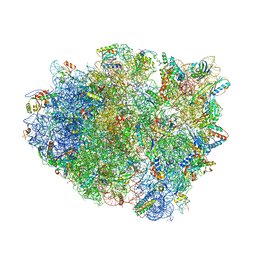 | | Crystal structure analysis of ribosomal decoding (near-cognate tRNA-tyr complex). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4V53
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with gentamicin. | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 16S rRNA, 23S rRNA, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4V6D
 
 | | Crystal structure of the E. coli 70S ribosome in an intermediate state of ratcheting | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, W, Dunkle, J.A, Cate, J.H.D. | | Deposit date: | 2009-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.814 Å) | | Cite: | Structures of the ribosome in intermediate States of ratcheting.
Science, 325, 2009
|
|
