2KRX
 
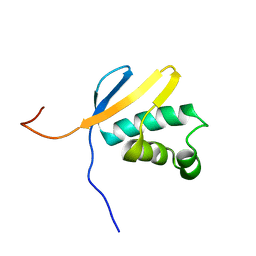 | | Solution NMR Structure of asl3597 from Nostoc sp. PCC7120. Northeast Structural Genomics Consortium Target ID Nsr244. | | Descriptor: | Asl3597 protein | | Authors: | Feldmann, E.A, Ramelot, T.A, Yang, Y, Lee, D.K, Ciccosanti, C, Janjua, H.A, Liu, J, Rost, B, Acton, T, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-12-22 | | Release date: | 2010-03-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Asl3597 from Nostoc sp. PCC7120, the first structure from protein domain family PF12095, reveals a novel fold.
Proteins, 80, 2012
|
|
2KFP
 
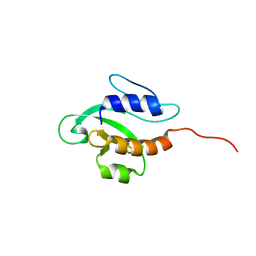 | | Solution NMR structure of PSPTO_3016 from Pseudomonas syringae. Northeast Structural Genomics Consortium target PsR293. | | Descriptor: | PSPTO_3016 protein | | Authors: | Feldmann, E.A, Ramelot, T.A, Zhao, L, Hamilton, K, Ciccosanti, C, Xiao, R, Nair, R, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR and X-ray crystal structures of Pseudomonas syringae Pspto_3016 from protein domain family PF04237 (DUF419) adopt a "double wing" DNA binding motif.
J.Struct.Funct.Genom., 13, 2012
|
|
2LNU
 
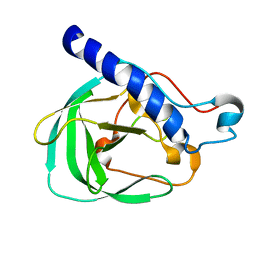 | | Solution NMR Structure of the uncharacterized protein from gene locus rrnAC0354 of Haloarcula marismortui, Northeast Structural Genomics Consortium Target HmR11 | | Descriptor: | Uncharacterized protein | | Authors: | Rossi, P, Liu, G, Lange, O.F, Lee, H, Janjua, H, Ciccosanti, C, Wang, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-05 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2KAD
 
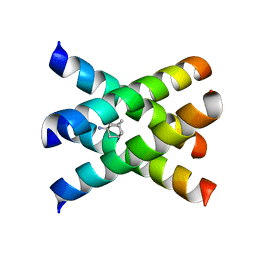 | | Magic-Angle-Spinning Solid-State NMR Structure of Influenza A M2 Transmembrane Domain | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, Transmembrane peptide of Matrix protein 2 | | Authors: | Hong, M, Cady, S.D, Mishanina, T.V. | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Structure of amantadine-bound M2 transmembrane peptide of influenza A in lipid bilayers from magic-angle-spinning solid-state NMR: the role of Ser31 in amantadine binding.
J.Mol.Biol., 385, 2009
|
|
2KKI
 
 | |
2M99
 
 | |
1WQE
 
 | | An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis | | Descriptor: | OmTx3 | | Authors: | Chagot, B, Pimentel, C, Dai, L, Pil, J, Tytgat, J, Nakajima, T, Corzo, G, Darbon, H, Ferrat, G. | | Deposit date: | 2004-09-28 | | Release date: | 2005-01-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis
Biochem.J., 388, 2005
|
|
1WQC
 
 | | An unusual fold for potassium channel blockers : NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis | | Descriptor: | OmTx1 | | Authors: | Chagot, B, Pimentel, C, Dai, L, Pil, J, Tytgat, J, Nakajima, T, Corzo, G, Darbon, H, Ferrat, G. | | Deposit date: | 2004-09-27 | | Release date: | 2005-01-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis
Biochem.J., 388, 2005
|
|
2FY9
 
 | |
2HM9
 
 | | Solution structure of dihydrofolate reductase complexed with trimethoprim, 33 structures | | Descriptor: | 2,4-DIAMINO-5-(3,4,5-TRIMETHOXY-BENZYL)-PYRIMIDIN-1-IUM, Dihydrofolate reductase | | Authors: | Polshakov, V.I, Birdsall, B. | | Deposit date: | 2006-07-11 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structures of apo L.casei dihydrofolate reductase and its complexes with trimethoprim and NADPH. Contributions to positive cooperative binding from ligand-induced refolding, conformational changes and interligand hydrophobic interactions
Biochemistry, 2011
|
|
2LM4
 
 | | Solution NMR Structure of mitochondrial succinate dehydrogenase assembly factor 2 from Saccharomyces cerevisiae, Northeast Structural Genomics Consortium Target YT682A | | Descriptor: | Succinate dehydrogenase assembly factor 2, mitochondrial | | Authors: | Eletsky, A, Winge, D.R, Lee, H, Lee, D, Kohan, E, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2011-11-22 | | Release date: | 2012-01-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of yeast succinate dehydrogenase flavinylation factor sdh5 reveals a putative sdh1 binding site.
Biochemistry, 51, 2012
|
|
2KZN
 
 | | Solution NMR Structure of Peptide methionine sulfoxide reductase msrB from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR10 | | Descriptor: | Peptide methionine sulfoxide reductase msrB | | Authors: | Ertekin, A, Maglaqui, M, Janjua, H, Cooper, B, Ciccosanti, C, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J, Lee, H, Aramini, J.M, Rossi, P, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LSY
 
 | |
1W09
 
 | | Solution structure of the cis form of the human alpha-hemoglobin stabilizing protein (AHSP) | | Descriptor: | ALPHA-HEMOGLOBIN STABILIZING PROTEIN | | Authors: | Santiveri, C.M, Perez-Canadillas, J.M, Vadivelu, M.K, Allen, M.D, Rutherford, T.J, Watkins, N.A, Bycroft, M. | | Deposit date: | 2004-05-25 | | Release date: | 2004-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the alpha-hemoglobin stabilizing protein: insights into conformational heterogeneity and binding.
J. Biol. Chem., 279, 2004
|
|
2JZ4
 
 | | Putative 32 kDa myrosinase binding protein At3g16450.1 from Arabidopsis thaliana | | Descriptor: | Jasmonate inducible protein isolog | | Authors: | Takeda, N, Sugimori, N, Torizawa, T, Terauchi, T, Ono, A.M, Yagi, H, Yamaguchi, Y, Kato, K, Ikeya, T, Guntert, P, Aceti, D.J, Markley, J.L, Kainosho, M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-12-28 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the putative 32 kDa myrosinase-binding protein from Arabidopsis (At3g16450.1) determined by SAIL-NMR.
Febs J., 275, 2008
|
|
1WQD
 
 | | An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis | | Descriptor: | OmTx2 | | Authors: | Chagot, B, Pimentel, C, Dai, L, Pil, J, Tytgat, J, Nakajima, T, Corzo, G, Darbon, H, Ferrat, G. | | Deposit date: | 2004-09-28 | | Release date: | 2005-01-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis
Biochem.J., 388, 2005
|
|
1XWU
 
 | | Solution structure of ACAUAGA loop | | Descriptor: | 5'-R(*CP*GP*AP*AP*AP*CP*AP*UP*AP*GP*AP*UP*UP*CP*GP*A)-3' | | Authors: | Sakamoto, T, Oguro, A, Kawai, G, Ohtsu, T, Nakamura, Y. | | Deposit date: | 2004-11-02 | | Release date: | 2005-02-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structures of double loops of an RNA aptamer against mammalian initiation factor 4A
Nucleic Acids Res., 33, 2005
|
|
2GM0
 
 | | Linear dimer of stemloop SL1 from HIV-1 | | Descriptor: | RNA (35-MER) | | Authors: | Ulyanov, N.B, Mujeeb, A, Du, Z, Tonelli, M, Parslow, T.G, James, T.L. | | Deposit date: | 2006-04-05 | | Release date: | 2006-04-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Full-length Linear Dimer of Stem-Loop-1 RNA in the HIV-1 Dimer Initiation Site.
J.Biol.Chem., 281, 2006
|
|
2K7Y
 
 | |
2KDZ
 
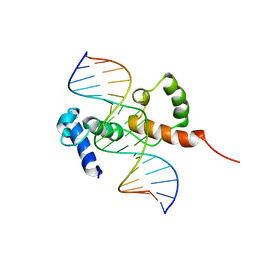 | | Structure of the R2R3 DNA binding domain of MYB1 protein from protozoan parasite trichomonas vaginalis in complex with MRE-1/MRE-2R DNA | | Descriptor: | 5'-D(*AP*AP*GP*AP*TP*AP*AP*CP*GP*AP*TP*AP*TP*TP*TP*A)-3', 5'-D(*TP*AP*AP*AP*TP*AP*TP*CP*GP*TP*TP*AP*TP*CP*TP*T)-3', MYB24 | | Authors: | Lou, Y.C, Wei, S.Y, Rajasekaran, M, Chou, C.C, Hsu, H.M, Tai, J.H, Chen, C. | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of DNA recognition by a novel Myb1 DNA-binding domain in the protozoan parasite Trichomonas vaginalis.
Nucleic Acids Res., 2009
|
|
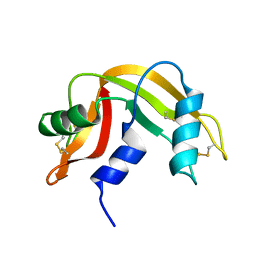
2LFJ
 
 | |
2LP6
 
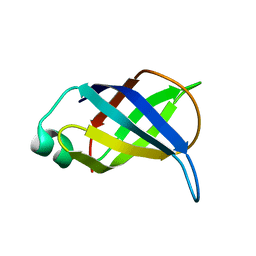 | | Refined Solution NMR Structure of the 50S ribosomal protein L35Ae from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target (NESG) PfR48 | | Descriptor: | 50S ribosomal protein L35Ae | | Authors: | Snyder, D.A, Aramini, J.M, Yu, B, Huang, Y.J, Xiao, R, Cort, J.R, Shastry, R, Ma, L, Liu, J, Rost, B, Acton, T.B, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-02-02 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the ribosomal protein RP-L35Ae from Pyrococcus furiosus.
Proteins, 80, 2012
|
|
203D
 
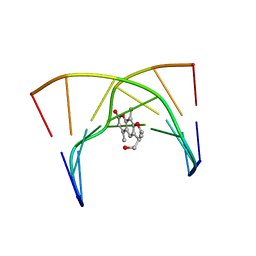 | | THE SOLUTION STRUCTURES OF PSORALEN MONOADDUCTED AND CROSSLINKED DNA OLIGOMERS BY NMR SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | 4'-HYDROXYMETHYL-4,5',8-TRIMETHYLPSORALEN, DNA (5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3') | | Authors: | Spielmann, H.P, Dwyer, T.J, Hearst, J.E, Wemmer, D.E. | | Deposit date: | 1995-04-06 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of psoralen monoadducted and cross-linked DNA oligomers by NMR spectroscopy and restrained molecular dynamics.
Biochemistry, 34, 1995
|
|
204D
 
 | | THE SOLUTION STRUCTURES OF PSORALEN MONOADDUCTED AND CROSSLINKED DNA OLIGOMERS BY NMR SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | 4'-HYDROXYMETHYL-4,5',8-TRIMETHYLPSORALEN, DNA (5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3') | | Authors: | Spielmann, H.P, Dwyer, T.J, Hearst, J.E, Wemmer, D.E. | | Deposit date: | 1995-04-06 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of psoralen monoadducted and cross-linked DNA oligomers by NMR spectroscopy and restrained molecular dynamics.
Biochemistry, 34, 1995
|
|
2L15
 
 | | Solution Structure of Cold Shock Protein CspA Using Combined NMR and CS-Rosetta method | | Descriptor: | Cold shock protein CspA | | Authors: | Tang, Y, Schneider, W.M, Shen, Y, Raman, S, Inouye, M, Baker, D, Roth, M.J, Montelione, G.T. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Fully automated high-quality NMR structure determination of small (2)H-enriched proteins.
J Struct Funct Genomics, 11, 2010
|
|
